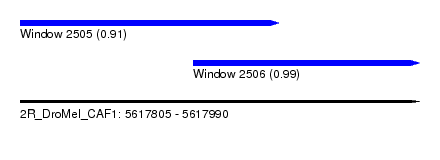
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,617,805 – 5,617,990 |
| Length | 185 |
| Max. P | 0.987986 |

| Location | 5,617,805 – 5,617,925 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5617805 120 + 20766785 GGGUAAAAUGCAGCCAACACAUCCCAUCCGUUUGGGUCUGGCCCUUAACUUCUCAGUCUUCUACUAUGAGAUUUUGAACUCACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAA .(((((...((............(((......)))((((((...((((..(((((...........)))))..)))).....))))))...))))))).(((((((.....))))))).. ( -27.70) >DroVir_CAF1 12297 120 + 1 GGGCAAAAUGCAGCCAACACAUCCCAUCCGCUUGGGUCUUGCACUUAACUUCUCAGUCUUCUAUUAUGAGAUUUUGAAUUCUCCAGAUAAGGCUUGCCAAUUGGCUAAACAGGUUAAUAU .((((......((((....((.((((......))))...))...((((..(((((...........)))))..)))).............)))))))).(((((((.....))))))).. ( -25.80) >DroSec_CAF1 7470 120 + 1 GGGUAAAAUGCAGCCAACACAUCCCAUCCGUUUGGGUCUGGCCCUUAACUUCUCAGUCUUCUACUAUGAGAUUUUGAACUCACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAA .(((((...((............(((......)))((((((...((((..(((((...........)))))..)))).....))))))...))))))).(((((((.....))))))).. ( -27.70) >DroSim_CAF1 9098 120 + 1 GGGUAAAAUGCAGCCAACACAUCCCAUCCGUUUGGGUCUGGCCCUUAACUUCUCAGUCUUCUACUAUGAGAUUUUGAACUCACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAA .(((((...((............(((......)))((((((...((((..(((((...........)))))..)))).....))))))...))))))).(((((((.....))))))).. ( -27.70) >DroAna_CAF1 8008 120 + 1 GGGUAAAAUGCAGCCAACACAUCCCAUCCGUUUGGGUCUGGCCCUCAACUUCUCAGUCUUCUACUAUGAGAUUUUGAACUCGCCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAA .(((((...((............(((......)))(((((((..((((..(((((...........)))))..))))....)))))))...))))))).(((((((.....))))))).. ( -33.60) >consensus GGGUAAAAUGCAGCCAACACAUCCCAUCCGUUUGGGUCUGGCCCUUAACUUCUCAGUCUUCUACUAUGAGAUUUUGAACUCACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAA .(((((...((............(((......)))((((((...((((..(((((...........)))))..)))).....))))))...))))))).(((((((.....))))))).. (-27.32 = -27.04 + -0.28)


| Location | 5,617,885 – 5,617,990 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.06 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5617885 105 + 20766785 CACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAACA-CACU---GAGACUUGGCUCGAUUAAUC---GAAAUGUAUGGCAAGCGAGCAAGAAAGGCACACAAAAUC-------- ...........((((((((....(((((.(((((.......)-).))---)....)))))((((....))---))......))))))))..((.......))..........-------- ( -23.00) >DroVir_CAF1 12377 93 + 1 CUCCAGAUAAGGCUUGCCAAUUGGCUAAACAGGUUAAUAUUC-AAGU---G------------------C---CAAUUGUCUGGCAGGCGAGC--AAGAGUCAAGCCAAGUGCAACAACA .....(((...(((((((.(((((((.....)))))))....-...(---(------------------(---((......))))))))))))--....)))..((.....))....... ( -26.10) >DroSec_CAF1 7550 103 + 1 CACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAACA-GACU---GAGAUUUGGCUCGAUUAAUC---GAAAUGUAUGGCAAGCGAGC--GAAAGGCACACAAAAUU-------- ...........((((((((....((((((..((((.......-))))---....))))))((((....))---))......))))))))..((--.....))..........-------- ( -25.20) >DroSim_CAF1 9178 103 + 1 CACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAACA-CACU---GAGACUUGGCUCGAUUAAUC---GAAAUGUAUGGCAAGCGAGC--GAAAGGCACACAAAAUU-------- ...........((((((((....(((((.(((((.......)-).))---)....)))))((((....))---))......))))))))..((--.....))..........-------- ( -23.50) >DroAna_CAF1 8088 101 + 1 CGCCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAAUAAUACUAUCAAGGCU---AUCCAUUAAUUUGAGAAAUGUAUGGCAAGCA--------AGGCACAAAAAACA-------- .(((.......(((((((.(((((((.....)))))))....((((..(((((..(---(.....)).))))).....))))))))))).--------.)))..........-------- ( -22.80) >consensus CACCAGACAAAGCUUGCCAAUUGGCUAAACAGGUUAAUAACA_CACU___GAGACUUGGCUCGAUUAAUC___GAAAUGUAUGGCAAGCGAGC__GAAAGGCACACAAAAUU________ ...........(((((((((((((((.....)))))))............(((......)))...................))))))))............................... (-16.92 = -17.46 + 0.54)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:39 2006