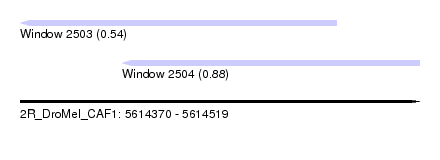
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,614,370 – 5,614,519 |
| Length | 149 |
| Max. P | 0.876949 |

| Location | 5,614,370 – 5,614,488 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.37 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
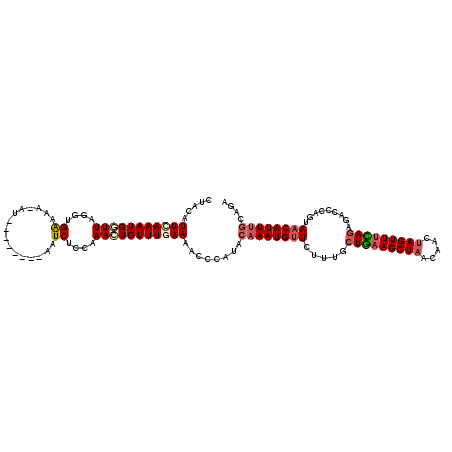
>2R_DroMel_CAF1 5614370 118 - 20766785 UCCAAAAUGUUUGAAACCCCAUACAAAUGUUCUUUACUGAAGCUAACAACUAGUUUCAAAGACCCAGUAACAUUGGCAGAAGCAAUUAUAAAUGCUGAUUUCUGCGCAGUUGCCUAAA .......(((.((......)).))).....((((...((((((((.....))))))))))))....(((((..(((((((((((........))))....))))).)))))))..... ( -23.40) >DroSec_CAF1 3952 117 - 1 UCCAAACUGUUUGAAAAC-CAUACAAAUGUUGUUUGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGAAGCAAUUAUAAAUGCCGAGUUCUGCGCAGUUGUCUGAA ....((((((........-.....(((((((((((.(((((((((.....))))))))))))).....)))))))(((((((((........)))....))))))))))))....... ( -30.00) >DroSim_CAF1 3976 118 - 1 UCCAAACUGUUUGAAAACCCAUACAAAUGUUGUACGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGAAGCAAUUAUAAAUGCUGAGUUCUGCGCAGUUGUCUAAA ....((((((..............(((((((((...(((((((((.....)))))))))..)).....)))))))(((((((((........))))....)))))))))))....... ( -29.40) >DroEre_CAF1 4154 118 - 1 UCAAAAUUGUUUCAAAACCUAUACAAAUGUUCUUCUCUGCAGCUUACAACUAGUUUUAGAUACCCAGUAACAUUUGCAGAAGCAAUUAUAAAUACUGAGUUCUGCGCAGUUGUCUAAA ....(((((((((..........((((((((((..((((.((((.......)))).)))).....)).))))))))..)))))))))......((((.(.....).))))........ ( -18.80) >DroYak_CAF1 3954 111 - 1 UAAAAAUUGUUUGAAAACCCAUAGAAAUGGUCUUUGCUAAAGCUAAAAACUAGUUUUAGAGACCCAGUAACAUUUGCACAAGCAAUUAU-------GAGUUCUGCGCAGUUGUCUAAA ....((((((.....(((.(((((....(((((...(((((((((.....)))))))))))))).........((((....))))))))-------).)))....))))))....... ( -23.90) >consensus UCCAAACUGUUUGAAAACCCAUACAAAUGUUCUUUGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGAAGCAAUUAUAAAUGCUGAGUUCUGCGCAGUUGUCUAAA ....((((((..............(((((((.....(((((((((.....))))))))).........)))))))((((((..................))))))))))))....... (-16.99 = -17.83 + 0.84)

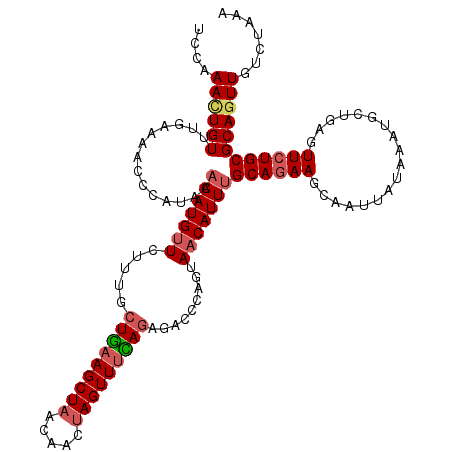
| Location | 5,614,408 – 5,614,519 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -12.71 |
| Energy contribution | -13.91 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5614408 111 - 20766785 CUACAUUGAAAUGGUUAGGUGAAAA-AU--------AGUCUCCAAAAUGUUUGAAACCCCAUACAAAUGUUCUUUACUGAAGCUAACAACUAGUUUCAAAGACCCAGUAACAUUGGCAGA ..(((((...((((...(((...((-((--------(..........)))))...)))))))...)))))((((...((((((((.....)))))))))))).(((((...))))).... ( -20.30) >DroSec_CAF1 3990 110 - 1 CUACAUUCAAAUGGUUAUGUGAAAA-AU--------AAUCUCCAAACUGUUUGAAAAC-CAUACAAAUGUUGUUUGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGA .....(((((((((((.((.((...-..--------....)))))))))))))))...-....((((((((((((.(((((((((.....))))))))))))).....)))))))).... ( -27.50) >DroSim_CAF1 4014 111 - 1 CUACAUUCAAAUGGUUAUGUGAAAA-AU--------AAUCUCCAAACUGUUUGAAAACCCAUACAAAUGUUGUACGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGA .....(((((((((((.((.((...-..--------....)))))))))))))))........((((((((((...(((((((((.....)))))))))..)).....)))))))).... ( -26.20) >DroEre_CAF1 4192 119 - 1 UUACUUUUAAAUGUUUAAAUGAACAGAU-AAAAGCUUCUCUCAAAAUUGUUUCAAAACCUAUACAAAUGUUCUUCUCUGCAGCUUACAACUAGUUUUAGAUACCCAGUAACAUUUGCAGA ...((((((..(((((....)))))..)-))))).............................((((((((((..((((.((((.......)))).)))).....)).)))))))).... ( -17.50) >DroYak_CAF1 3985 120 - 1 UUACAUUUAAAUGAUUAGAUGGAGAGAUGAAAAGCUUCUCUAAAAAUUGUUUGAAAACCCAUAGAAAUGGUCUUUGCUAAAGCUAAAAACUAGUUUUAGAGACCCAGUAACAUUUGCACA ((((.((((((..(((...(((((((.(....)..)))))))..)))..)))))).............(((((...(((((((((.....))))))))))))))..)))).......... ( -27.60) >consensus CUACAUUCAAAUGGUUAGGUGAAAA_AU________AAUCUCCAAACUGUUUGAAAACCCAUACAAAUGUUCUUUGCUGAAGCUAACAACUAGUUUCAGAGACCCAGUAACAUUUGCAGA .....(((((((((((....((................))....)))))))))))........((((((((.....(((((((((.....))))))))).........)))))))).... (-12.71 = -13.91 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:37 2006