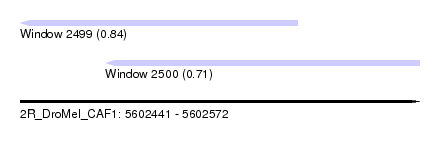
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,602,441 – 5,602,572 |
| Length | 131 |
| Max. P | 0.843362 |

| Location | 5,602,441 – 5,602,532 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -26.33 |
| Energy contribution | -28.53 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5602441 91 - 20766785 ACGGCCGCCUGCAGCGAAAAGGCGGCGGUGCGCUGUGGUGUCGCUAGGAUGCACUGCACCACCGCCAGCAGCACCAACAUGGCACCACUGG ...(((((((.........)))))))((((((((((((((..((.((......))))..))))).)))).)))))..((((....)).)). ( -42.40) >DroVir_CAF1 23 79 - 1 ACGGCCGCCUGCAGCGAGUAGGCGC---CUCGCUGGGGCGUCGCCAGGA--CAAUGCAGC----CGAGCAGGCCCAGGAUGGCAACA---G ...(((((((.(((((((.......---)))))))(((((((.....))--)..(((...----...))).))))))).))))....---. ( -34.50) >DroSec_CAF1 23 91 - 1 ACGGCCGCCUGCAGCGAAAAGGCGGCGGUGCGCUGUGGUGUCGCUAGGAUGCACUGCACCACCGUCAGCAGCACCACCAUGGCACCACUGG ...(((((((.........)))))))((((((((((((((..((.((......))))..))))).)))).))))).(((((....)).))) ( -44.40) >DroEre_CAF1 77 91 - 1 ACGGCCGCCUGCAGCGAAAAUGCGGCGGUGCGUUGUGGUGUCGCUAGGAUGCACUGCACCACCGCCAGCAGCACCAGCAUGGCACCACCGG (((((((((.(((.......))))))))).))).(((((((((((.((.(((.((((......).)))..))))))))..))))))))... ( -38.80) >DroYak_CAF1 79 91 - 1 ACGGCCGCCUGCAGCGAAAAUGCGGCGGUGCGUUGCGGUGUCGCUAGGAUGCACUGCAUCACCGCCAGCAGCACCACCAUGGCACCACUGG ...((((((.(((.......))))))((((((((((((((..((.((......))))..))))).)))).))))).....)))........ ( -40.70) >DroPer_CAF1 23 88 - 1 ACGGCUGCCUGGAGCGAAUAGGCGGCGGUGCGCUGUGGUGUUGCCCAGAUGCACUGCAGGAG---UAGUAGCACCACGAAGCUACCGCAAA ...((((((((.......))))))))(((((..(((((((((((.....(((...)))....---..)))))))))))..)).)))..... ( -38.70) >consensus ACGGCCGCCUGCAGCGAAAAGGCGGCGGUGCGCUGUGGUGUCGCUAGGAUGCACUGCACCACCGCCAGCAGCACCACCAUGGCACCACUGG ...(((((((.........)))))))((((((((((((((..((.((......))))..))))).)))).)))))................ (-26.33 = -28.53 + 2.20)



| Location | 5,602,469 – 5,602,572 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -45.85 |
| Consensus MFE | -25.95 |
| Energy contribution | -27.37 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5602469 103 - 20766785 CAGUGGAUACUUGGCCGCCUCCCGACGAUGCAGUUCAUCCACGGCCGCCUGCAGCGAAAAGGCGGCGGUGCGCUGUGGUGUCGCUAGGAUGCACUGC--ACCACC--- ..((((....((((.......))))...((((((.(((((...(((((((.........)))))))(((((((....))).)))).))))).)))))--))))).--- ( -44.60) >DroVir_CAF1 47 95 - 1 UAAUGGAUACUUGGCAGCCUCGCGGCGAUGCAGCUCAUCGACGGCCGCCUGCAGCGAGUAGGCGC---CUCGCUGGGGCGUCGCCAGGA--CAAUGC--AGC------ .........((((((.(((....)))(((((..((((.(((.(((.((((((.....))))))))---)))).))))))))))))))).--......--...------ ( -43.90) >DroSec_CAF1 51 103 - 1 CAGUGGAUACUUGGCCGCCUCGCGACGAUGCAUUUCAUCCACGGCCGCCUGCAGCGAAAAGGCGGCGGUGCGCUGUGGUGUCGCUAGGAUGCACUGC--ACCACC--- (((((.((.((((((((((.((((..((((.....))))(((.(((((((.........))))))).)))))).).))))..)))))))).))))).--......--- ( -45.50) >DroMoj_CAF1 48 97 - 1 CAAUGGAAACUUGGCAGCCUCGCGGCGAUGCAGCUCAUCGACGGCCGCCUGCAGUGAGUAGGCGG---CGCGUUGGGGCGUGGCCAGGA--CAAUGCCAAGC------ .........(((((((.(((...(((.((((......(((((((((((((((.....))))))))---).)))))).)))).)))))).--...))))))).------ ( -51.00) >DroAna_CAF1 54 106 - 1 CAGCGGGUACUUGGCCGCCUCCCGACGAUGCAGUUCGUCCACAGCCGCCUGCAGCGAAUAGGCAGCGGUGCGUUGGGGCGUCGCCAGGAUGCACUGA--ACCACCAUC (((.(.((.(((((((((((...(((((......)))))..((((((((.((.((......)).)))))).)))))))))..)))))))).).))).--......... ( -42.40) >DroPer_CAF1 49 102 - 1 CAACGGAAAUUUCGAUGCCUCGCGACGGUGCAGCUCGUCAACGGCUGCCUGGAGCGAAUAGGCGGCGGUGCGCUGUGGUGUUGCCCAGAUGCACUGC--AGGAG---- ...((((...)))).((((((((..(((.(((((.((....))))))))))..))))...))))(((((((((((.((.....))))).))))))))--.....---- ( -47.70) >consensus CAAUGGAUACUUGGCCGCCUCGCGACGAUGCAGCUCAUCCACGGCCGCCUGCAGCGAAUAGGCGGCGGUGCGCUGGGGCGUCGCCAGGAUGCACUGC__ACCAC____ .........((((((.(((..(((..((((.....))))(((.((((((((.......)))))))).))))))...)))...)))))).................... (-25.95 = -27.37 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:33 2006