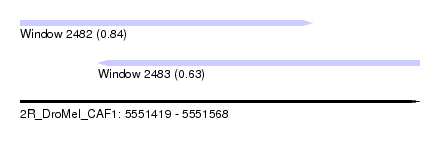
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,551,419 – 5,551,568 |
| Length | 149 |
| Max. P | 0.843705 |

| Location | 5,551,419 – 5,551,528 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
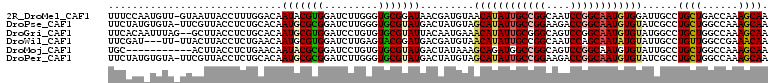
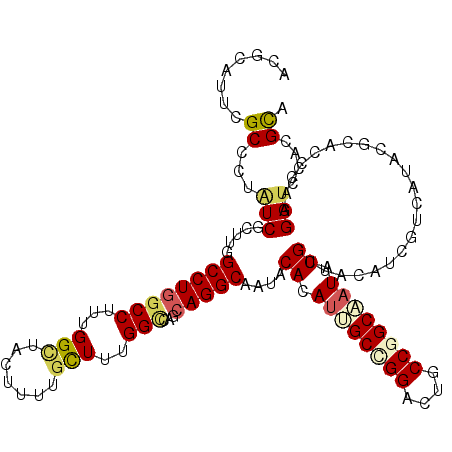
>2R_DroMel_CAF1 5551419 109 + 20766785 UUUCCAAUGUU-GUAAUUACCUUUGGACAAUACGUGGAUCUUGGGUGCGGAUAACGAUGUAACAUAUUGCCGGCAAUCCGGCAAUGUGGAUUGCCUGCUGACCAAAGCAA ..((((.((((-((............))))))..))))..((((..((((....((((....(((((((((((....))))))))))).)))).))))...))))..... ( -33.10) >DroPse_CAF1 5035 109 + 1 UUCUAUGUGUA-UUCGUUACCUCUGCACAAUGCGCGGAUCUUGGGUGCGUAUGACUAUGUAGCAUAUUGCCGGAAGACCGGCAAUGUGUAUCGCCUGCUGGCCAAAGCAA ...((((((((-((((.....(((((.......)))))...))))))))))))....(((.((((((((((((....))))))))))))...(((....)))....))). ( -37.60) >DroGri_CAF1 46176 108 + 1 UUCACAAUUUAG--GCUUACCUCUGCACAAUGCGUGGAUCCUGUGUGCGUAUUACAAUGAAACAUAUUGCGGGCAGUCCGGCAAUGUGUAUGGCCUGCUGGCCAAAGCAA ............--((((.....((((((.(((.((((..((((.((((..................)))).))))))))))).))))))(((((....))))))))).. ( -36.37) >DroWil_CAF1 2146 106 + 1 UUCGAU---UU-UUACUUACCUCUGAACAAUGCGUGGAUCUUGAGUACGGAUGACGAUGUAACAUAUUGCCGGCAAUCCAGCAAUAUGUAUUGCCUGUUGGCCGAAACAA ((((((---(.-.((((((..(((..((.....)))))...))))))..))).........(((((((((.((....)).)))))))))...(((....))))))).... ( -24.10) >DroMoj_CAF1 56514 99 + 1 UGC-----------ACUUACCUCUGAACAAUACGCGGAUCCUGUGUGCGUAUGACUAUAAAGCAGAUGGCCGGCAGUCCGGCAAUGUGUAUUGCCUGCUGGCCAAAGCAA (((-----------.......((((....(((((((.((...)).)))))))..........))))(((((((((....((((((....)))))))))))))))..))). ( -35.94) >DroPer_CAF1 4984 109 + 1 UUCUAUGUGUA-UUCGUUACCUCUGCACAAUGCGCGGAUCUUGGGUGCGUAUGACUAUGUAGCAUAUUGCCGGAAGACCGGCAAUGUGUAUCGCCUGCUGGCCAAAGCAA ...((((((((-((((.....(((((.......)))))...))))))))))))....(((.((((((((((((....))))))))))))...(((....)))....))). ( -37.60) >consensus UUCUAU_UGUA_UUACUUACCUCUGCACAAUGCGCGGAUCUUGGGUGCGUAUGACUAUGUAACAUAUUGCCGGCAGUCCGGCAAUGUGUAUUGCCUGCUGGCCAAAGCAA .............................(((((((.........))))))).........((((((((((((....))))))))))))......((((......)))). (-22.88 = -23.02 + 0.14)
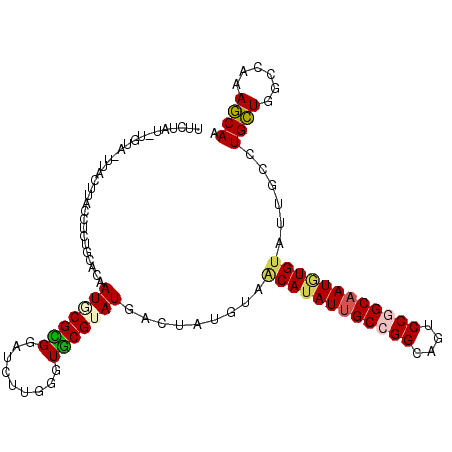
| Location | 5,551,448 – 5,551,568 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5551448 120 - 20766785 ACACAUUCGCCCUAUCGAUUGGCCUGGCCUUCGGCUACUUUUGCUUUGGUCAGCAGGCAAUCCACAUUGCCGGAUUGCCGGCAAUAUGUUACAUCGUUAUCCGCACCCAAGAUCCACGUA ........((......(((((.(((((((...(((.......)))..))))))..).)))))((.((((((((....)))))))).))..............))................ ( -33.10) >DroVir_CAF1 68701 120 - 1 ACGCUUUCGCCAUGUCCUUGGGCCUGGCCUUUGGCUACUUUUGCUUUGGCCAGCAGGCCAUACACAUAGCCGGUUUGCCGGCCUUAUGCUAUAUCGUGAUACGCACCCAGGAUCCGCGCA .......(((...(((((.((((.(((((((((((((.........))))))).))))))....(((((((((....)))))..))))..............)).)).)))))..))).. ( -43.40) >DroGri_CAF1 46204 120 - 1 ACGUCUUCGCCUUAUCGCUGGGCCUGGCCUUUGGCUACUUUUGCUUUGGCCAGCAGGCCAUACACAUUGCCGGACUGCCCGCAAUAUGUUUCAUUGUAAUACGCACACAGGAUCCACGCA ................(((((((((((((((((((((.........))))))).)))))..(((.(((((.((....)).))))).)))...................))).)))).)). ( -36.70) >DroWil_CAF1 2172 120 - 1 ACGUGUUUGCCCUAUCACUUGGCCUGACCUUUGGUUACUUUUGUUUCGGCCAACAGGCAAUACAUAUUGCUGGAUUGCCGGCAAUAUGUUACAUCGUCAUCCGUACUCAAGAUCCACGCA .((((.(((((.......((((((.((((...))))((....))...))))))..))))).((((((((((((....)))))))))))).........................)))).. ( -34.60) >DroMoj_CAF1 56533 120 - 1 AUGUCUUUGCCCUUUCGCUUGGCCUGGCCUUUGGCUACUUUUGCUUUGGCCAGCAGGCAAUACACAUUGCCGGACUGCCGGCCAUCUGCUUUAUAGUCAUACGCACACAGGAUCCGCGUA ...............(((....((((((...((((((.....((..(((((.(((((((((....))))))....))).)))))...))....))))))...))...))))....))).. ( -38.00) >DroAna_CAF1 4966 120 - 1 ACACAUUCGCUUUGUCGAUCGGCCUCGCCUUUGGAUACUUUUGUUUCGGUCCCCAGGCCAUUCACAUCGCAGGACUUCCGGCGAUAUGCUACAUUGUUAUUCGAACCCAGGAUCCCCGCA ........((...(..(((((((...)))((((((((.(..(((..(((((....))))......(((((.((....)).)))))..)..)))..).)))))))).....))))..))). ( -28.00) >consensus ACGCAUUCGCCCUAUCGCUUGGCCUGGCCUUUGGCUACUUUUGCUUUGGCCAGCAGGCAAUACACAUUGCCGGACUGCCGGCAAUAUGCUACAUCGUCAUACGCACCCAGGAUCCACGCA ........((...(((.....((((((((...(((.......)))..)))...)))))....((.((((((((....)))))))).))......................)))....)). (-23.14 = -23.50 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:16 2006