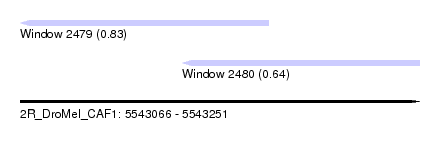
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,543,066 – 5,543,251 |
| Length | 185 |
| Max. P | 0.825979 |

| Location | 5,543,066 – 5,543,181 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -18.97 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
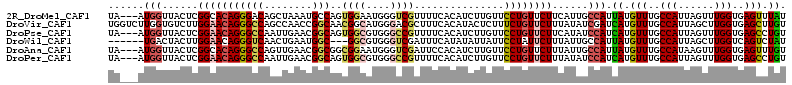
>2R_DroMel_CAF1 5543066 115 - 20766785 UUCAUUGCCAUUAUGUUUGCCAUUAGUUUGGUGAGUUUAUUUGGCUACCACAUCUACCUUGUGCUGGUGAAUCGCACGACAUUGGGUAAGUACCUAAAGCCUGACUUA--UAAAUCA .(((..((((.((..((..(((......)))..))..))..)))).........(((((.(((.(.(((.....))).)))).))))).............)))....--....... ( -23.90) >DroVir_CAF1 29635 114 - 1 UUUAUAUCGAUCAUGUUUGCCAUUAGCUUGGUGAGCUUGUUCGGCUACCACAUCUACCUGGUGCUCGUCAAUCGCACCACAUUGGGUAAGCGA-GAAACUCUCACUUUA-UAUACC- .......(((.((.(((..(((......)))..))).)).)))((((((.((......((((((.........))))))...))))).)))((-(.....)))......-......- ( -32.60) >DroPse_CAF1 5638 115 - 1 UUCAUAUCCAUCAUGUUUGCCAUUAGUUUGGUGAGCCUGUUUGGCUAUCACAUCUAUUUGGUUCUGGUCAAUCGCACCACUUUAGGUGAGUAG--AGAACCGGGCGUGAUUAAACUA .........(((((((..((((.(((..((((.((((.....))))))))...)))..)))).(((((...((.((((......))))....)--)..))))))))))))....... ( -32.80) >DroYak_CAF1 4373 115 - 1 UUUAUUGCUAUUAUGUUUGCCAUUAGUUUGGUGAGUUUGUUUGGCUACCACAUCUACCUUGUGCUAGUGAAUCGCACGACAUUGGGUAAGUACUUAAAGCCGGACUUA--UAAUUUA .........((((((((..(((......)))..))...((((((((.......((((((.(((...(((.....)))..))).)))).)).......)))))))).))--))))... ( -27.14) >DroMoj_CAF1 5705 106 - 1 UUCAUAUCGAUCAUGUUUGCCAUUAGCUUGGUGAGCUUAUUUGGAUACCACAUCUAUCUGGUGCUCGUCAACCGCACCACACUGGGUAAU------AUCUGUGA---AA-UAUACC- ((((((((((....(((..(((......)))..)))....)))).((((.((......((((((.........))))))...))))))..------...)))))---).-......- ( -26.80) >DroPer_CAF1 5677 115 - 1 UUUAUAUCCAUCAUGUUUGCCAUUAGUUUGGUGAGCCUGUUUGGCUAUCACAUCUAUUUGGUUCUGGUCAAUCGCACCACUUUAGGUGAGUAG--AGAACCGGGCGUGAUUAAACUA .........(((((((..((((.(((..((((.((((.....))))))))...)))..)))).(((((...((.((((......))))....)--)..))))))))))))....... ( -32.80) >consensus UUCAUAUCCAUCAUGUUUGCCAUUAGUUUGGUGAGCUUGUUUGGCUACCACAUCUACCUGGUGCUGGUCAAUCGCACCACAUUGGGUAAGUAC__AAAACCGGACUUAA_UAAACCA .............((((((((..(((..((((.((((.....))))))))...)))..((((((.........)))))).....))))))))......................... (-18.97 = -18.42 + -0.55)
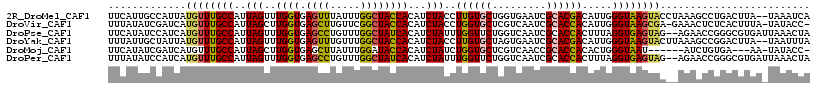
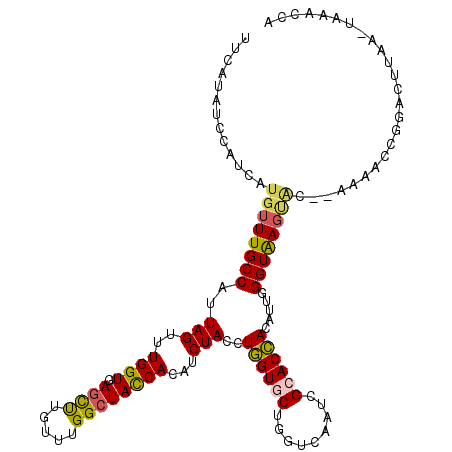
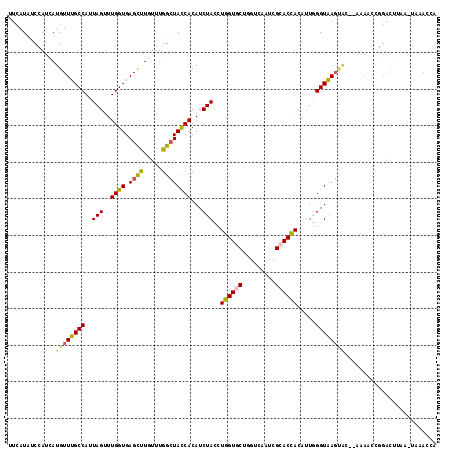
| Location | 5,543,141 – 5,543,251 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
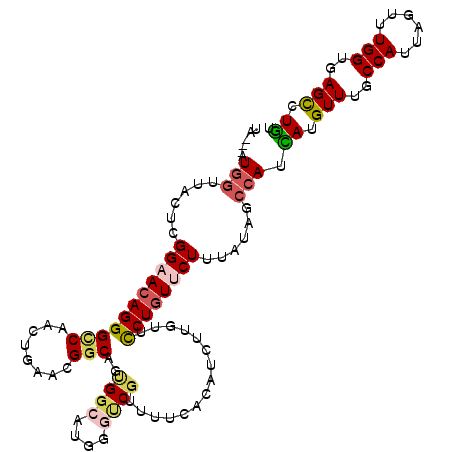
>2R_DroMel_CAF1 5543141 110 - 20766785 UA---AUGGUUACUCGGCACAGGGACAGCUAAAUGCCAGUGGAAUGGGUCGUUUUCACAUCUUGUUCCUGUUCUUCAUUGCCAUUAUGUUUGCCAUUAGUUUGGUGAGUUUAU ((---(((((.....((.(((((((((((.....))..((((((........))))))....))))))))).)).....)))))))..((..(((......)))..))..... ( -33.90) >DroVir_CAF1 29709 113 - 1 UGGUCUUGGUGUCUUGGAACAGGGCCAGCCAACCGGCAACGGCAUGGGACGCUUUCACAUACUCUUUCUGUUCUUUAUAUCGAUCAUGUUUGCCAUUAGCUUGGUGAGCUUGU (((((..(((((...(((((((((((.(((....)))...)))((((((....))).)))......))))))))..)))))))))).(((..(((......)))..))).... ( -34.40) >DroPse_CAF1 5713 110 - 1 UA---AUGGUUACUCGGAACAGGGCCAAUUGAACGGCAGUGGCGUGGGCCGUUUUCACAUCUUGUUCCUGUUCUUCAUAUCCAUCAUGUUUGCCAUUAGUUUGGUGAGCCUGU ..---((((......((((((((..(((..(((((((..........))))))).......)))..))))))))......))))((.(((..(((......)))..))).)). ( -35.90) >DroWil_CAF1 12129 104 - 1 ------UGACUACUUGGAACAGGGUCAACUGAAUGGC---GGCGUGGGUCGAUUUCAUAUAUUAUUCCUAUUCUUUAUUGCCAUUAUGUUUGCCAUUAGCUUGGUCAGUCUAU ------.(((((((.((.....(((.(((..((((((---((.(((((..(((.......)))...))))).)).....))))))..))).))).....)).))).))))... ( -25.20) >DroAna_CAF1 5056 110 - 1 UA---AUGGUUACUCGGCACAGGGCCAGUUGAACGGCGGCGGAAUGGGUCGAUUCCACAUCUUGUUCCUGUUCUUUAUUGCCAUUAUGUUUGCCAUAAGUUUGGUGAGUUUGU ((---(((((.(((.(((.....)))))).((((((..(((..(((((......)).)))..)))..))))))......)))))))..((..(((......)))..))..... ( -32.50) >DroPer_CAF1 5752 110 - 1 UA---AUGGUUACUCGGAACAGGGCCAAUUGAACGGCAGUGGCGUGGGCCGUUUUCACAUCUUGUUCCUGUUCUUUAUAUCCAUCAUGUUUGCCAUUAGUUUGGUGAGCCUGU ..---((((......((((((((..(((..(((((((..........))))))).......)))..))))))))......))))((.(((..(((......)))..))).)). ( -35.90) >consensus UA___AUGGUUACUCGGAACAGGGCCAACUGAACGGCAGUGGCAUGGGUCGUUUUCACAUCUUGUUCCUGUUCUUUAUAGCCAUCAUGUUUGCCAUUAGUUUGGUGAGCCUGU ......(((......(((((((((((........)))..((((....))))...............))))))))......))).((.(((..(((......)))..))).)). (-20.20 = -20.57 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:13 2006