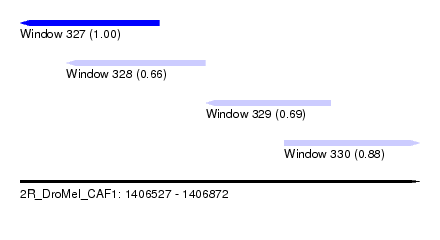
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,406,527 – 1,406,872 |
| Length | 345 |
| Max. P | 0.999583 |

| Location | 1,406,527 – 1,406,647 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -35.71 |
| Energy contribution | -35.75 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1406527 120 - 20766785 UUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCAGCAGCAUACUGUUAGAAUCCACUUGGGGAGCUCUUAAGUG ......((((.((((((((...........)))).))))(..((((((((((...((((((((((((.......))))).......)))))))..)))))).))))..)))))....... ( -37.61) >DroSec_CAF1 32333 120 - 1 UUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCAGCAGCAUACUGUUAGAAUCCACUUGGGGAGCUUUUAAGUG (((((((((.(((((((((...........)))).)))))......((((((...((((((((((((.......))))).......)))))))..))))))))))))))).......... ( -35.11) >DroSim_CAF1 37344 120 - 1 UUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCAGAAGCAUACUGUUAGAAUCCACUUGGGGAGCUCUUAAGUG ......((((.((((((((...........)))).))))(..((((((((((...((((((((((((.......))))).......)))))))..)))))).))))..)))))....... ( -37.61) >DroEre_CAF1 30845 119 - 1 UUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGA-GCCGCAGCACCAUACUGUUAGAAUCCACUUGGGGAGCUCUUAAGUG ......((((.((((((((...........)))).))))(..((((((((((...((((((((((((......-))))).......)))))))..)))))).))))..)))))....... ( -38.11) >DroYak_CAF1 32794 120 - 1 UUCUUGGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCACAGCAGCAUACUGUUAGAAUCCACUUGGGGAGCUCUUAAGUG ..((((((((.((((((((...........)))).))))(..((((((((((...((((((((((....)....((....)).)).)))))))..)))))).))))..))).)))))).. ( -37.30) >consensus UUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCAGCAGCAUACUGUUAGAAUCCACUUGGGGAGCUCUUAAGUG ......((((.((((((((...........)))).))))(..((((((((((...((((((((((((.......))))).......)))))))..)))))).))))..)))))....... (-35.71 = -35.75 + 0.04)

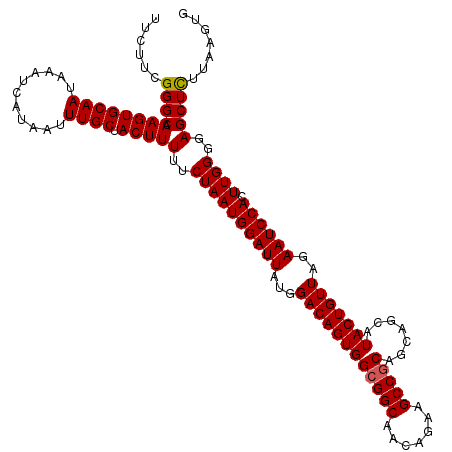
| Location | 1,406,567 – 1,406,687 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1406567 120 - 20766785 CCAGCGCUCGCACACUUUGUGGCUCUCAUGAAUUUUUCCAUUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCA ....((((.((.(((...)))))((.(((((.....((((((....(((.(((((((((...........)))).))))).)))))))))))))))).))))(((((.......))))). ( -30.50) >DroSec_CAF1 32373 120 - 1 GCAGCGCUCGCACACUUUGCGGCUCUCAUGAAUUUUUCCAUUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCA ...((((.((((.....))))))((.(((((.....((((((....(((.(((((((((...........)))).))))).))))))))))))))))..)).(((((.......))))). ( -31.90) >DroSim_CAF1 37384 120 - 1 CCAGCGCUCGCACACUUUGCGGCUCUCAUGAAUUUUUCCAUUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCA (((..((.((((.....)))))).............((((((....(((.(((((((((...........)))).))))).)))))))))...)))......(((((.......))))). ( -31.90) >DroEre_CAF1 30885 119 - 1 CCAGCGCUCGCACACUUUGCGGCUCUCAUGAAUUUUUCCAUUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGA-GCCGCA ...((....))......((((((((((((((.....((((((....(((.(((((((((...........)))).))))).)))))))))))))).........(....))))-)))))) ( -33.50) >DroYak_CAF1 32834 120 - 1 CCAGCGCUCGCACACUUUGUGGCUCUCAUGAAUUUUUCCAUUCUUGGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCACA ...(((((..(((.....)))((((.((.((((......)))).))))))..)))))......(((((((((..((.......))..)))))))))...(((((..(....)..))))). ( -31.10) >consensus CCAGCGCUCGCACACUUUGCGGCUCUCAUGAAUUUUUCCAUUCUUCGGGCAAAGUGCAAUAAAUCAUAAUUUGCUACUUUUUCUAAUGGAUUAUGGACAGUGGCGGCAACAGAAGCCGCA ...((((.((((.....))))))((.(((((.....((((((....(((.(((((((((...........)))).))))).))))))))))))))))..)).(((((.......))))). (-29.90 = -29.86 + -0.04)

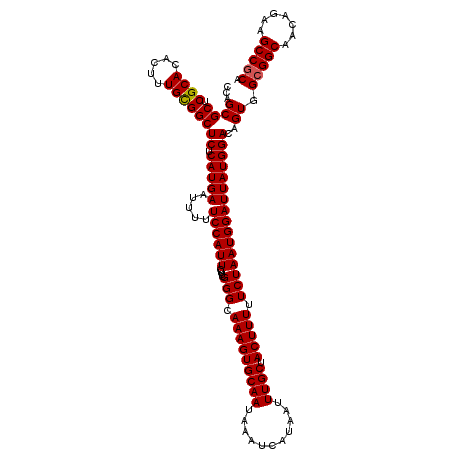
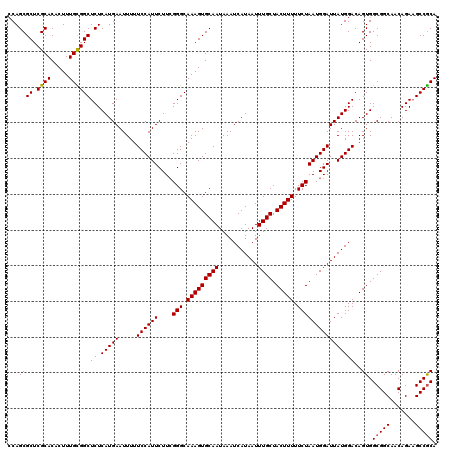
| Location | 1,406,687 – 1,406,795 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1406687 108 - 20766785 UUGGUAUGCCACUUUGGAGUUUUUGCCUUCACGACCCCAUGCCCCGUCCUUCUCCAC----------CCCCAUCAC-CCUUCCAUACC-AUCCUGGCUUCAUUUUAGCGGAGCAUACUUU .(((((((......(((((.......))))).(((..........))).........----------.........-.....))))))-).....(((((........)))))....... ( -16.80) >DroSec_CAF1 32493 109 - 1 UUUGUAUGCCACUUUGGAGUUUUUGCCUCCACGACCCCAUGCCCCGUCCUCCGCCAC----------CCCCACCGCCCCUUCCAUGCC-AUCCUGGCUUCAUUUCAGCGGAGCAUACUUU ...((((((.....(((((.......)))))..........................----------.....((((.........(((-.....))).........)))).))))))... ( -24.07) >DroSim_CAF1 37504 119 - 1 UUGGUAUGCCACUUUGGAGUUUUUGCCUCCACGACCCCAUGCCCCGUCCUCCGCCACCCGGGUGCACCCCCACCGCCCCUUCCAUUCC-AUCCUGGCUUCAUUUCAGCGGAGCAUACUUU ..((((((......(((((.......)))))......))))))..((.((((((.....(((.((.........)))))..(((....-....)))..........)))))).))..... ( -29.90) >DroEre_CAF1 31004 96 - 1 UUCGUAUGCCACUUUGGAGUUUUUGCCCC-----C--------UAGAGCUGCCCGCG----------GUCCCUG-CCCCUCGCAUCGCAGUGCUGGCUUCCAUUCAGCGGAGCAUACUUU ...((((((..(.(((((......(((..-----.--------.((.(((((..(((----------(......-....))))...))))).))))).....))))).)..))))))... ( -27.20) >DroYak_CAF1 32954 101 - 1 UUCGUAUGCCACAUUGGAGUUUUUGCCUCUACUGC--------CUGUACUUCCCCAU----------GCCCUUU-CCCCUCCCUUUGCAGUCCUGGCUUCAAUUCAGCGGAGCAUACUUU ...((((((..(.(((((......(((...(((((--------..............----------.......-...........)))))...))).....))))).)..))))))... ( -22.25) >consensus UUCGUAUGCCACUUUGGAGUUUUUGCCUCCACGACCCCAUGCCCCGUCCUCCCCCAC__________CCCCAUC_CCCCUUCCAUUCC_AUCCUGGCUUCAUUUCAGCGGAGCAUACUUU ...((((((..(.(((((((....(((...................................................................)))...))))))).)..))))))... (-14.03 = -14.47 + 0.44)


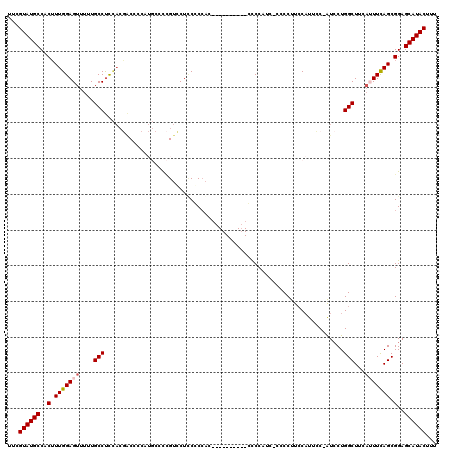
| Location | 1,406,755 – 1,406,872 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -11.94 |
| Energy contribution | -13.58 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1406755 117 + 20766785 AUGGGGUCGUGAAGGCAAAAACUCCAAAGUGGCAUACCAAAAUGUAAAACUCUUGUAAUACCCUAGCCACUGU---GGGCCCCUCCAACCGAUGAUUUACCCAGCUUUGGGGCCACCAAA .((((((..((....))...)))))).((((((.(((......)))..((....)).........))))))..---.((((((.........................))))))...... ( -29.31) >DroSec_CAF1 32562 120 + 1 AUGGGGUCGUGGAGGCAAAAACUCCAAAGUGGCAUACAAAAAUGUAAAACUCUUGUAAUACCCCAGCCACUGUGCUGGGCCACUCCAACCGCUGAUUUAACCAGCUUUGGGGCCACCAAA .(((((((.(((((.......))))).((((((.((((....))))................(((((......)))))))))))((((..((((.......)))).)))))))).))).. ( -43.20) >DroSim_CAF1 37583 120 + 1 AUGGGGUCGUGGAGGCAAAAACUCCAAAGUGGCAUACCAAAAUGUAAAACUCUUGUAAUACCCCAGCCAAUGUGCUGGGCCACUCCAACCGCUGAUUUACCCAGCUUUGGGGCCCCCAAA ..((((((.(((((.......))))).((((((..........(((..((....))..))).(((((......)))))))))))((((..((((.......)))).)))))))))).... ( -44.30) >DroEre_CAF1 31070 102 + 1 -----G-----GGGGCAAAAACUCCAAAGUGGCAUACGAAAAUGUAAAACUCUUGUAAUACCCCGGCCAGCGU---GGCCCACUCCACCCAG---UUCACCCAGCUUUGGGCC--CCCAA -----(-----(((((.......(((((((((...((......(((..((....))..)))...((((.....---))))...........)---)....))).)))))))))--))).. ( -31.01) >DroYak_CAF1 33020 96 + 1 -----GCAGUAGAGGCAAAAACUCCAAUGUGGCAUACGAAAAUGUAAAACUCUUGUAAUACCCUGGCCACUGU---GGCCCACUCCACCCAC-----CAAUCAA-----------CCAAA -----......(((.......)))....((((..(((((.............))))).......((((.....---))))........))))-----.......-----------..... ( -14.02) >consensus AUGGGGUCGUGGAGGCAAAAACUCCAAAGUGGCAUACCAAAAUGUAAAACUCUUGUAAUACCCCAGCCACUGU___GGGCCACUCCAACCGCUGAUUUACCCAGCUUUGGGGCC_CCAAA .............((......((((((((((((.(((......)))..((....)).........)))........((......)).................)))))))))...))... (-11.94 = -13.58 + 1.64)

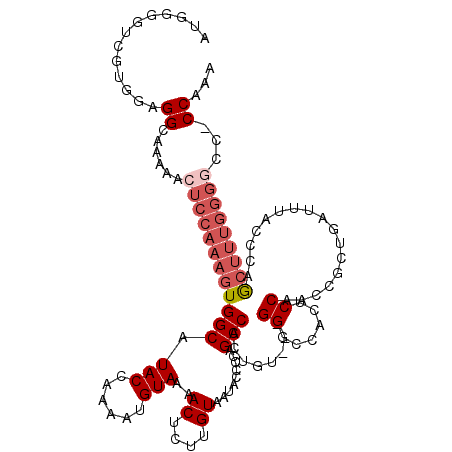
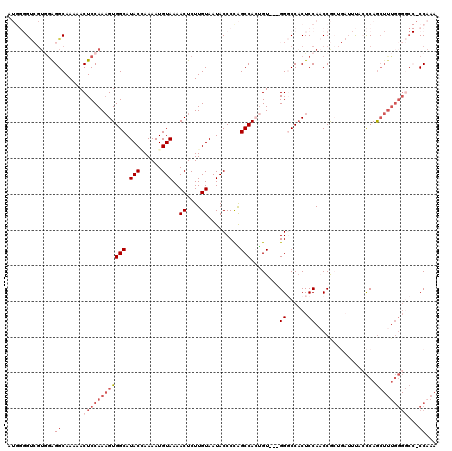
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:02 2006