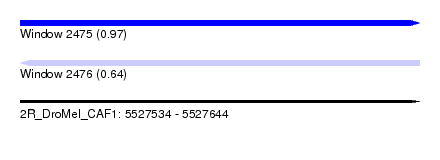
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,527,534 – 5,527,644 |
| Length | 110 |
| Max. P | 0.973680 |

| Location | 5,527,534 – 5,527,644 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.08 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5527534 110 + 20766785 ------AGAAUGACAUU-UGCU--UUAUUAUGCCAGCCAACAGUGUCCAACUUACGGAACCCAACCUAUACUUCCAAGUGGCAGGAAGUGAGGUUGACAAGGCGAACAAGGCUGUU-GUA ------......(((..-.(((--((.((.((((..((...(((.....)))...))....((((((.(((((((........)))))))))))))....)))))).)))))...)-)). ( -32.00) >DroSec_CAF1 50822 118 + 1 AACUCAAGAAUGGCAUUUUUCC--UUAUUAUGCCAGCAAACAGUUUCCAUCUUACGGAACUCAACCUGUACUUCCAAGUGGCAGGAAGUGAGGUUGACAAGGCGAACAAGGCUGUUGGUA ..........((((((..(...--..)..))))))((.((((((((((.......)))).(((((((.(((((((........)))))))))))))).............)))))).)). ( -34.90) >DroSim_CAF1 28130 112 + 1 ------AGAAUGGCAUUUUUCC--UUAUUAUGCCAGCAAACAGUUUCCAUCUUACGGAACUCAACCUGUACUUCCAAGUGGCAGGAAGUGAGGUUGACAAGGCGAACAAGGCUGUUGGUA ------....((((((..(...--..)..))))))((.((((((((((.......)))).(((((((.(((((((........)))))))))))))).............)))))).)). ( -34.90) >DroEre_CAF1 27908 112 + 1 AACCAA------GCAUUUACCCAAUUUUUUUUUCAACAAACAGUAUCCAACUUAUGGAACGCAGCCUGUACUUCCAAGUGGCAGGAAGUGAGGUUGACAA-GCGAACAAGGCUGUU-AUA ......------..........................((((((.((((.....))))..(((((((.(((((((........))))))))))))).)..-.........))))))-... ( -26.90) >DroYak_CAF1 28968 97 + 1 AACUAA------AAAAU----------------AUAGAAAUGGUAUCCAACUUAUGGAACUCGGCCUGUACUUCCAAGUGGCAGGAAGUGAGGUUGACAAGGCGAACAAGGCUGUU-AUA ......------.....----------------.....((((((.((((.....))))..(((((((.(((((((........)))))))))))))).............))))))-... ( -23.80) >consensus AAC__AAGAAUGGCAUUUUUCC__UUAUUAUGCCAGCAAACAGUAUCCAACUUACGGAACUCAACCUGUACUUCCAAGUGGCAGGAAGUGAGGUUGACAAGGCGAACAAGGCUGUU_GUA ...................................((.((((((.(((.......)))...((((((.(((((((........)))))))))))))..............)))))).)). (-25.08 = -24.52 + -0.56)

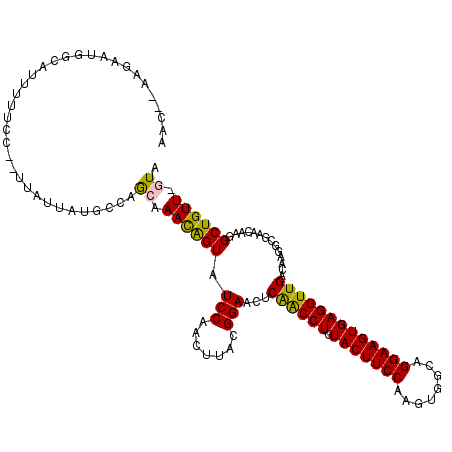
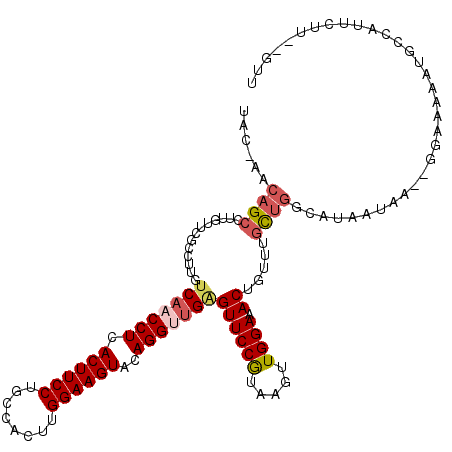
| Location | 5,527,534 – 5,527,644 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -19.78 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5527534 110 - 20766785 UAC-AACAGCCUUGUUCGCCUUGUCAACCUCACUUCCUGCCACUUGGAAGUAUAGGUUGGGUUCCGUAAGUUGGACACUGUUGGCUGGCAUAAUAA--AGCA-AAUGUCAUUCU------ ..(-(((((...((((((.(((((((((((.((((((........))))))..)))))))......)))).))))))))))))..((((((.....--....-.))))))....------ ( -32.70) >DroSec_CAF1 50822 118 - 1 UACCAACAGCCUUGUUCGCCUUGUCAACCUCACUUCCUGCCACUUGGAAGUACAGGUUGAGUUCCGUAAGAUGGAAACUGUUUGCUGGCAUAAUAA--GGAAAAAUGCCAUUCUUGAGUU .........(((((((.(((...(((((((.((((((........))))))..))))))).....(((((..(....)..))))).)))..)))))--)).................... ( -32.80) >DroSim_CAF1 28130 112 - 1 UACCAACAGCCUUGUUCGCCUUGUCAACCUCACUUCCUGCCACUUGGAAGUACAGGUUGAGUUCCGUAAGAUGGAAACUGUUUGCUGGCAUAAUAA--GGAAAAAUGCCAUUCU------ .........(((((((.(((...(((((((.((((((........))))))..))))))).....(((((..(....)..))))).)))..)))))--))..............------ ( -32.80) >DroEre_CAF1 27908 112 - 1 UAU-AACAGCCUUGUUCGC-UUGUCAACCUCACUUCCUGCCACUUGGAAGUACAGGCUGCGUUCCAUAAGUUGGAUACUGUUUGUUGAAAAAAAAAUUGGGUAAAUGC------UUGGUU ...-..((((...((..((-((((.......((((((........)))))))))))).))((((((.....)))).)).....)))).......(((..(((....))------)..))) ( -25.41) >DroYak_CAF1 28968 97 - 1 UAU-AACAGCCUUGUUCGCCUUGUCAACCUCACUUCCUGCCACUUGGAAGUACAGGCCGAGUUCCAUAAGUUGGAUACCAUUUCUAU----------------AUUUU------UUAGUU ...-(((.......((((.(((((.......((((((........))))))))))).)))).((((.....))))............----------------.....------...))) ( -17.71) >consensus UAC_AACAGCCUUGUUCGCCUUGUCAACCUCACUUCCUGCCACUUGGAAGUACAGGUUGAGUUCCGUAAGUUGGAAACUGUUUGCUGGCAUAAUAA__GGAAAAAUGCCAUUCUU__GUU ......((((.............(((((((.((((((........))))))..)))))))((((((.....)))).)).....))))................................. (-19.78 = -20.58 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:09 2006