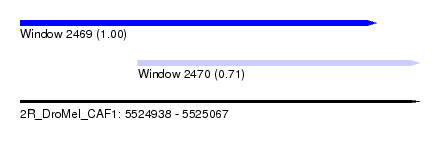
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,524,938 – 5,525,067 |
| Length | 129 |
| Max. P | 0.995724 |

| Location | 5,524,938 – 5,525,053 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -29.77 |
| Energy contribution | -30.18 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5524938 115 + 20766785 CUGGAUGGAUGCUUAAAUUGAUGUCCAUUAGCGUUGAAUUAGUGAGCAAUUAUGUUGCCUGCUCGCUAGGGCGGGCAGCAGAUACAGAUACUCGUCUAUAGAUGGAGAUUCAGAU ((((((...((((((..(..((((......))))..).....))))))....((((((((((((....))))))))))))..........((((((....))).))))))))).. ( -39.60) >DroSec_CAF1 48224 115 + 1 CUGGAUGGAUGCUUAAAUUGAUGUCCAUUAGCGUUAAAUUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAU ((((((...((((((..(((((((......))))))).....))))))....((((((((((((....))))))))))))..(((((((....))))))).......)))))).. ( -42.50) >DroSim_CAF1 25524 115 + 1 CUGGAUGGAUGCUUAAAUUGAUGUCCAUUAGCGUUAAAUUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAU ((((((...((((((..(((((((......))))))).....))))))....((((((((((((....))))))))))))..(((((((....))))))).......)))))).. ( -42.50) >DroEre_CAF1 25389 89 + 1 CUGGAUGGAUGCUUAAAUUGAUGUCCAUUAGUGUUAAAUUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAG--------------------------AU ...(((((((((.......).)))))))).(((((......((((....))))(((((((((((....))))))))))).)))))..--------------------------.. ( -31.70) >DroYak_CAF1 26639 114 + 1 CUGGAUGGAUGCUGAAAUUGAUGUCCAUUAGCGUUAAAUUAGUGAGCAAUUAUGUUACCUGCUCGCUGGGGCG-GCAGCAGAUACAGAUACUUGUCUGCAGAUGGAGAUUCAGAU ((((((...(((((...(((((((......))))))).((((((((((...........))))))))))..))-)))(((((((........)))))))........)))))).. ( -36.30) >consensus CUGGAUGGAUGCUUAAAUUGAUGUCCAUUAGCGUUAAAUUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAU (((......((((((..(((((((......))))))).....))))))....((((((((((((....))))))))))))....)))....(((((....))))).......... (-29.77 = -30.18 + 0.41)

| Location | 5,524,976 – 5,525,067 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5524976 91 + 20766785 UUAGUGAGCAAUUAUGUUGCCUGCUCGCUAGGGCGGGCAGCAGAUACAGAUACUCGUCUAUAGAUGGAGAUUCAGAUACUUUUAAAGGUGG ....((((......((((((((((((....))))))))))))..........((((((....))).))).))))..(((((....))))). ( -27.80) >DroSec_CAF1 48262 91 + 1 UUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAUACUUUUAAAGGUGG ....(((((.....((((((((((((....))))))))))))..(((((((....)))))))......).))))..(((((....))))). ( -30.70) >DroSim_CAF1 25562 91 + 1 UUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAUACUUUUAAAGGUGG ....(((((.....((((((((((((....))))))))))))..(((((((....)))))))......).))))..(((((....))))). ( -30.70) >DroYak_CAF1 26677 90 + 1 UUAGUGAGCAAUUAUGUUACCUGCUCGCUGGGGCG-GCAGCAGAUACAGAUACUUGUCUGCAGAUGGAGAUUCAGAUACUUUUAAAGGUGG ((((((((((...........))))))))))....-...(((((((........)))))))...............(((((....))))). ( -21.80) >consensus UUAGUGAGCAAUUAUGUUGCCUGCUCGCUGGGGCGGGCAGCAGAUACAGAUACUCGUCUGUAGAUGGAGAUUCAGAUACUUUUAAAGGUGG ....(((((.....((((((((((((....))))))))))))..(((((((....)))))))......).))))..(((((....))))). (-25.23 = -26.23 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:03 2006