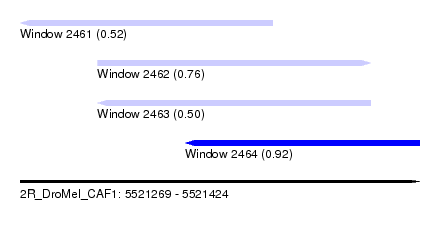
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,521,269 – 5,521,424 |
| Length | 155 |
| Max. P | 0.924544 |

| Location | 5,521,269 – 5,521,367 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5521269 98 - 20766785 CACCACCACCUGCAUCUAU----GUACACCUGCGGCUGCAGUAAAAAUGACAUUAAAAUGAAAGUCAUAU-----GCACAUGGUUAACUCGCCA-CGCAUAACCAAGC .........(((((....(----(((....))))..)))))......((((.((......)).))))(((-----((...((((......))))-.)))))....... ( -16.60) >DroSec_CAF1 44425 102 - 1 CACCACCACCUGCUCCUAUGUAUGUCCACCUGCAGCUGCAGUAAAAGUGACAUUAAAAUGGAAGUCAUAU-----GCACAUGGUUAACUCGCCA-CGCAUAACCAAGU ...........(((.((((..(((((.((((((....)))).....)))))))....)))).)))..(((-----((...((((......))))-.)))))....... ( -20.70) >DroSim_CAF1 21716 102 - 1 CACCACCACCUGCACCUAUGUAUGUACACCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAU-----GCACAUGGUUAACUCGCCA-CGCAUAACCAAGC .........(((((....((((.(....).))))..)))))......((((.((......)).))))(((-----((...((((......))))-.)))))....... ( -18.30) >DroEre_CAF1 21453 82 - 1 CACCACCACCUGCA--------------A------CUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAU-----GCACAUGGUUAACUCGCUA-CGCAUAACCAAGC .........((((.--------------.------..))))......((((.((......)).))))(((-----((...((((......))))-.)))))....... ( -12.60) >DroWil_CAF1 28161 91 - 1 ------GUCCUGCA-----------GCACCUGCAGUUGCAGUAAAAAUGGCAUUAAAAUGAAAGUCAUAUAUGCAGCACAUGGUUAACUUUUCAGCGCAUAACUAAGC ------...(((((-----------((.......))))))).....(((((.((......)).))))).(((((.((....((....)).....)))))))....... ( -18.00) >DroYak_CAF1 21938 88 - 1 CACCACCACCUGCA--------------GCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAU-----GCACAUGGUUAACUCGCCA-CGCAUAACCAAGC .........(((((--------------((....)))))))......((((.((......)).))))(((-----((...((((......))))-.)))))....... ( -22.40) >consensus CACCACCACCUGCA____________CACCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAU_____GCACAUGGUUAACUCGCCA_CGCAUAACCAAGC ..........(((................((((....)))).....(((((.((......)).))))).......)))..((((((....((....)).))))))... (-13.01 = -13.07 + 0.06)

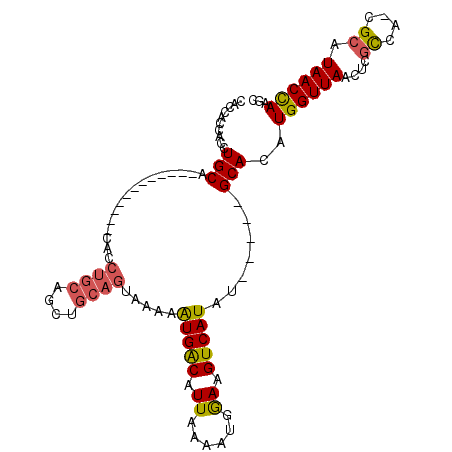

| Location | 5,521,299 – 5,521,405 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5521299 106 + 20766785 GCAUAUGACUUUCAUUUUAAUGUCAUUUUUACUGCAGCCGCAGGUGUAC----AUAGAUGCAGGUGGUGGUGAAAGGUGAAAGCACCUAACCGCACCCACCCACCGUCGA (((((((((............))))).....((((....))))......----....)))).(((((.((((..(((((....))))).....))))...)))))..... ( -32.30) >DroSec_CAF1 44455 110 + 1 GCAUAUGACUUCCAUUUUAAUGUCACUUUUACUGCAGCUGCAGGUGGACAUACAUAGGAGCAGGUGGUGGUGAAAGGUGAAGGCACCUAACCGCACCCACCCACCGUCGA .....((((((((......(((((.......((((....))))...))))).....))))..(((((.((((..(((((....))))).....))))...))))))))). ( -35.20) >DroSim_CAF1 21746 110 + 1 GCAUAUGACUUCCAUUUUAAUGUCAUUUUUACUGCAGCUGCAGGUGUACAUACAUAGGUGCAGGUGGUGGUGAAAGGUGAAGGCGCCUAACCGCACCCACCCACCGUCGA .....((((..........(((((((.....((((....))))))).)))).....((((..(((((..(((..(((((....)))))...)))..))))))))))))). ( -34.10) >DroEre_CAF1 21483 90 + 1 GCAUAUGACUUCCAUUUUAAUGUCAUUUUUACUGCAG------U--------------UGCAGGUGGUGGUGAAAGGUGAAAGCACCUAACCGCAUCCACCCAUCGUCAU ....(((((......................((((..------.--------------.))))(.(((((((..(((((....))))).....)).))))))...))))) ( -24.70) >DroYak_CAF1 21968 96 + 1 GCAUAUGACUUCCAUUUUAAUGUCAUUUUUACUGCAGCUGCAGC--------------UGCAGGUGGUGGUGAAAGGUGAAAGCACCUAACCGCGCCCACCCAUCGUCAC .....((((......................(((((((....))--------------)))))(.(((((((..(((((....))))).....)).))))))...)))). ( -31.30) >consensus GCAUAUGACUUCCAUUUUAAUGUCAUUUUUACUGCAGCUGCAGGUG_AC____AUAG_UGCAGGUGGUGGUGAAAGGUGAAAGCACCUAACCGCACCCACCCACCGUCGA (((((((((............))))).....((((....))))..............)))).(((((.((((..(((((....))))).....))))...)))))..... (-25.76 = -26.56 + 0.80)

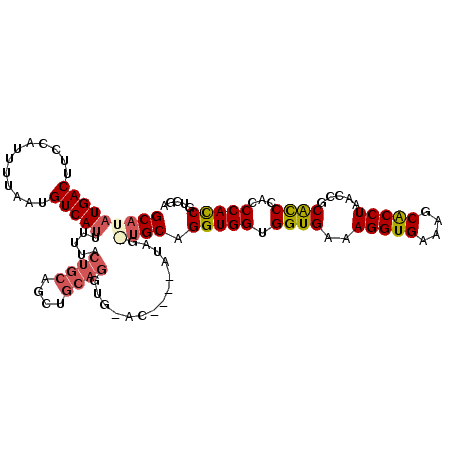
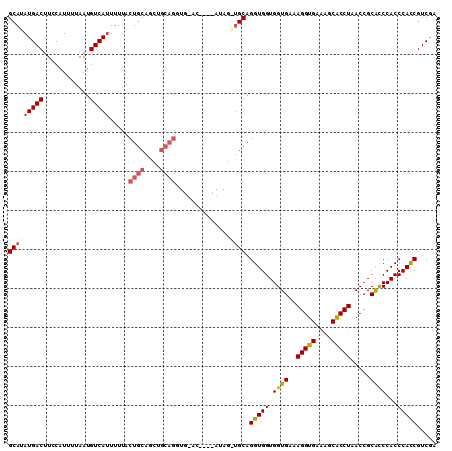
| Location | 5,521,299 – 5,521,405 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -23.38 |
| Energy contribution | -24.66 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5521299 106 - 20766785 UCGACGGUGGGUGGGUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCAUCUAU----GUACACCUGCGGCUGCAGUAAAAAUGACAUUAAAAUGAAAGUCAUAUGC ..(((.(..(((((.((..(..(((((....)))))..)..)))))))..)......----......((((....))))......................)))...... ( -30.30) >DroSec_CAF1 44455 110 - 1 UCGACGGUGGGUGGGUGCGGUUAGGUGCCUUCACCUUUCACCACCACCUGCUCCUAUGUAUGUCCACCUGCAGCUGCAGUAAAAGUGACAUUAAAAUGGAAGUCAUAUGC ..(((((..(((((.((..(..(((((....)))))..)..)))))))..)).((((..(((((.((((((....)))).....)))))))....))))..)))...... ( -37.70) >DroSim_CAF1 21746 110 - 1 UCGACGGUGGGUGGGUGCGGUUAGGCGCCUUCACCUUUCACCACCACCUGCACCUAUGUAUGUACACCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAUGC ..((((((((..(((((.(((.....)))..)))))....))))).((.......((((........((((....))))........))))......))..)))...... ( -31.61) >DroEre_CAF1 21483 90 - 1 AUGACGAUGGGUGGAUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA--------------A------CUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAUGC (((((...((((((.((..(..(((((....)))))..)..))))))))((.--------------.------..))........................))))).... ( -25.40) >DroYak_CAF1 21968 96 - 1 GUGACGAUGGGUGGGCGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA--------------GCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAUGC (((((....(((((.....(..(((((....)))))..).)))))..(((((--------------((....)))))))......................))))).... ( -32.90) >consensus UCGACGGUGGGUGGGUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA_CUAU____GU_CACCUGCAGCUGCAGUAAAAAUGACAUUAAAAUGGAAGUCAUAUGC ......((((((((.((..(..(((((....)))))..)..))))))))))................((((....)))).....(((((.((......)).))))).... (-23.38 = -24.66 + 1.28)

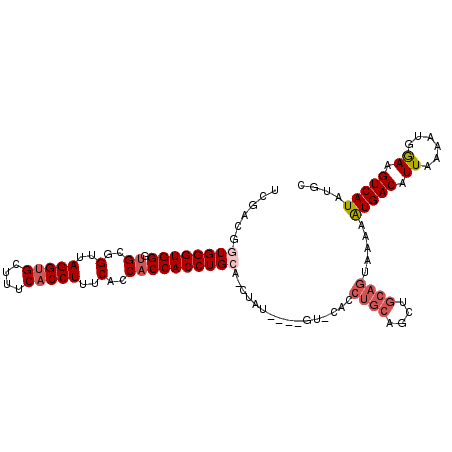
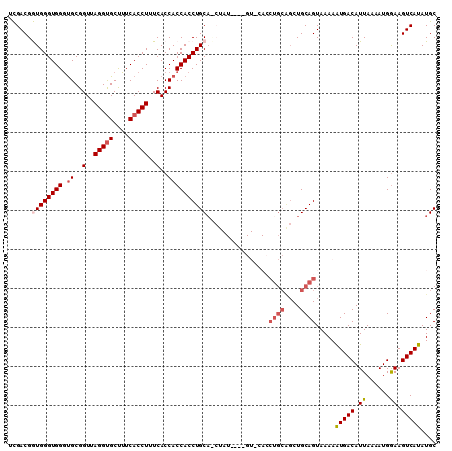
| Location | 5,521,333 – 5,521,424 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5521333 91 - 20766785 CCACUGUGGGUGGCCAACGUCGACGGUGGGUGGGUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCAUCUAU----GUACACCUGCGGCUG ...(((..((((..((.((....))(..(((((.((..(..(((((....)))))..)..)))))))..).....)----)..))))..)))... ( -38.20) >DroSec_CAF1 44489 95 - 1 CCACUGUGGGUGGCCAUCGUCGACGGUGGGUGGGUGCGGUUAGGUGCCUUCACCUUUCACCACCACCUGCUCCUAUGUAUGUCCACCUGCAGCUG ...(((..(((((.((((((.(..((..(((((.((..(..(((((....)))))..)..)))))))..)).).))).))).)))))..)))... ( -43.30) >DroSim_CAF1 21780 95 - 1 CCACUGUGGGUGGCCAUCGUCGACGGUGGGUGGGUGCGGUUAGGCGCCUUCACCUUUCACCACCACCUGCACCUAUGUAUGUACACCUGCAGCUG ...(((..((((.((((((....))))))(((((((((((.(((........))).........))).)))))))).......))))..)))... ( -37.70) >DroEre_CAF1 21517 75 - 1 CCACUGUGGGUGGCCAUGGAUGACGAUGGGUGGAUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA--------------A------CUG ....((..(((((...(((.(((....(((((((.(((......))).))))))).)))))))))))..))--------------.------... ( -29.50) >DroYak_CAF1 22002 81 - 1 CCACUGUGGGUGGCCAUGGGUGACGAUGGGUGGGCGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA--------------GCUGCAGCUG .(((((..(((((.....(((((....((((((((((......)))))..))))).))))).)))))..))--------------).))...... ( -35.80) >consensus CCACUGUGGGUGGCCAUCGUCGACGGUGGGUGGGUGCGGUUAGGUGCUUUCACCUUUCACCACCACCUGCA_CUAU____GU_CACCUGCAGCUG .....(..(((((((((((....))))))........(((.(((((....)))))...))).)))))..)......................... (-27.18 = -27.82 + 0.64)

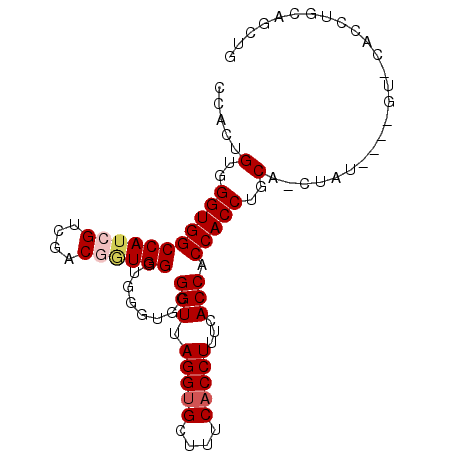

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:57 2006