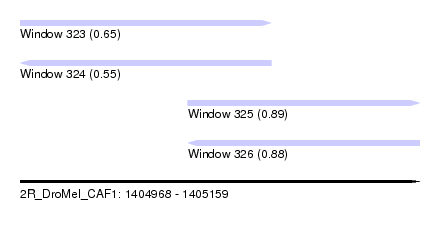
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,404,968 – 1,405,159 |
| Length | 191 |
| Max. P | 0.893374 |

| Location | 1,404,968 – 1,405,088 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -32.73 |
| Energy contribution | -32.40 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1404968 120 + 20766785 UCAGCUGGCUUCAGUUGGCGGUGUGAUGCCGAAGCCUCCUGACUUUGGGGUAGAGAACUGCAAGCUGCGGACUGCAAGCUGCGAACUGCAAACUGCAAACUACUAGCAGCUAGCUGCAAG .((((((((..(((((.(((((.((...(((.(((..(((......)))((((....))))..))).)))....)).))))).))))).....(((.........))))))))))).... ( -44.00) >DroSec_CAF1 30802 109 + 1 UCAGCUGCCUCCAGUUGGCG----GAUGCCGAAGCCUCCUCACUUCGGAGUAGAGAACUGCUAGCUGCGAACUGCAAACUGCAAACUGCAAACU-------GCUAGCUGCUAGCUGCUAG .((((((....(((((((((----(...((((((........))))))(((((....)))))...((((...(((.....)))...))))..))-------)))))))).)))))).... ( -38.40) >DroSim_CAF1 35811 109 + 1 UCAGCUGGCUCCAGUUGGCG----GAUGCCGAAGCCUCCUCACUUCGGAGUGGAGAACUGCUAGCUGCGAACUGCAAACUGCAAACUGCAAACU-------GCUAGCUGCUAGCUGCUAG .(((((((((((((((((((----(..((....))((((.(.(....).).))))..)))))))))).)).(((((...(((.....)))...)-------)).))..)))))))).... ( -42.90) >consensus UCAGCUGGCUCCAGUUGGCG____GAUGCCGAAGCCUCCUCACUUCGGAGUAGAGAACUGCUAGCUGCGAACUGCAAACUGCAAACUGCAAACU_______GCUAGCUGCUAGCUGCUAG .((((((((..(((((.((.........((((((........)))))).((((....)))).....)).))))).....(((.....)))..................)))))))).... (-32.73 = -32.40 + -0.33)

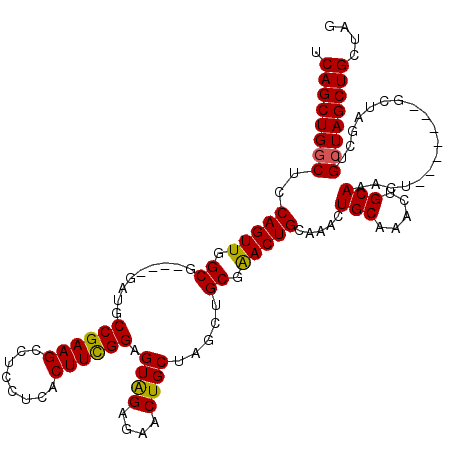
| Location | 1,404,968 – 1,405,088 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.25 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -32.05 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1404968 120 - 20766785 CUUGCAGCUAGCUGCUAGUAGUUUGCAGUUUGCAGUUCGCAGCUUGCAGUCCGCAGCUUGCAGUUCUCUACCCCAAAGUCAGGAGGCUUCGGCAUCACACCGCCAACUGAAGCCAGCUGA ....(((((((((((.(((.((((((((.((((.....)))).))))))...)).))).))))))...................(((((((((........)))....))))))))))). ( -40.20) >DroSec_CAF1 30802 109 - 1 CUAGCAGCUAGCAGCUAGC-------AGUUUGCAGUUUGCAGUUUGCAGUUCGCAGCUAGCAGUUCUCUACUCCGAAGUGAGGAGGCUUCGGCAUC----CGCCAACUGGAGGCAGCUGA ....(((((.((.((((((-------...((((.....))))..(((.....))))))))).........(((((...((.(((.((....)).))----)..))..)))))))))))). ( -42.90) >DroSim_CAF1 35811 109 - 1 CUAGCAGCUAGCAGCUAGC-------AGUUUGCAGUUUGCAGUUUGCAGUUCGCAGCUAGCAGUUCUCCACUCCGAAGUGAGGAGGCUUCGGCAUC----CGCCAACUGGAGCCAGCUGA .((((.((((((((((.((-------((.((((.....)))).))))))))....))))))...((..((((....))))..))(((((((((...----.)))....)))))).)))). ( -42.10) >consensus CUAGCAGCUAGCAGCUAGC_______AGUUUGCAGUUUGCAGUUUGCAGUUCGCAGCUAGCAGUUCUCUACUCCGAAGUGAGGAGGCUUCGGCAUC____CGCCAACUGGAGCCAGCUGA ....(((((....((((((..........((((.....))))..(((.....)))))))))...(((.(((......))).)))(((((((((........)))....))))))))))). (-32.05 = -32.50 + 0.45)

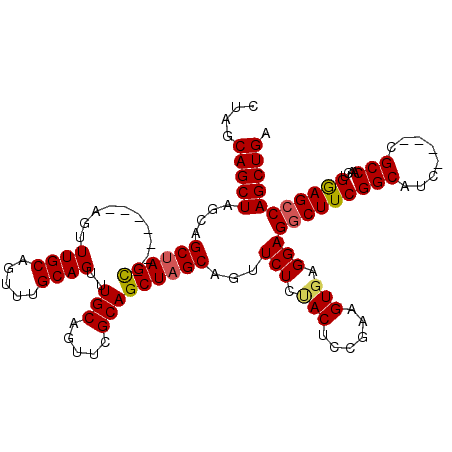
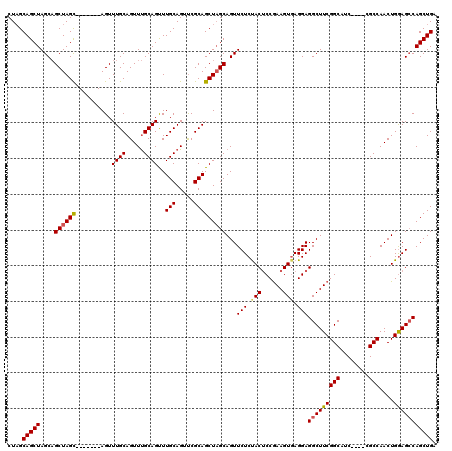
| Location | 1,405,048 – 1,405,159 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.91 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -31.93 |
| Energy contribution | -33.27 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1405048 111 + 20766785 GCGAACUGCAAACUGCAAACUACUAGCAGCUAGCUGCAAGC-UGCUAACUGCGAGUGGCAUCAGCGUCAUCGCCUGACGCUGACUGGUGGCCCUAACG---CACACAUUCAGCC-----A (((...(((.....)))......((((((((.......)))-)))))..)))...((((.(((((((((.....)))))))))...(((((......)---).))).....)))-----) ( -40.20) >DroSec_CAF1 30878 104 + 1 GCAAACUGCAAACU-------GCUAGCUGCUAGCUGCUAGCUUGCUAACUGCGAGUGGCAUCAGCGUCAUCGCUUGACGCUGACUGGUGGCCCCAACGACGCACGCA----GCC-----A ((.....))...((-------((..((..((((.(((((.(((((.....))))))))))(((((((((.....)))))))))))))..))......(.....))))----)..-----. ( -41.20) >DroSim_CAF1 35887 113 + 1 GCAAACUGCAAACU-------GCUAGCUGCUAGCUGCUAGCUUGCUAACUGCGAGUGGCAUCAGCGUCAUCGCCUGACGCGGACUGGUGGCCCCAACGACGCACACAUUCAGCCAGCCAA (((....((((...-------((((((........))))))))))....)))...((((....((((((.....))))))((.((((((((.........)).)))...))))).)))). ( -39.60) >consensus GCAAACUGCAAACU_______GCUAGCUGCUAGCUGCUAGCUUGCUAACUGCGAGUGGCAUCAGCGUCAUCGCCUGACGCUGACUGGUGGCCCCAACGACGCACACAUUCAGCC_____A ((.....))................((((((((.(((((.(((((.....))))))))))(((((((((.....)))))))))))))))))............................. (-31.93 = -33.27 + 1.33)

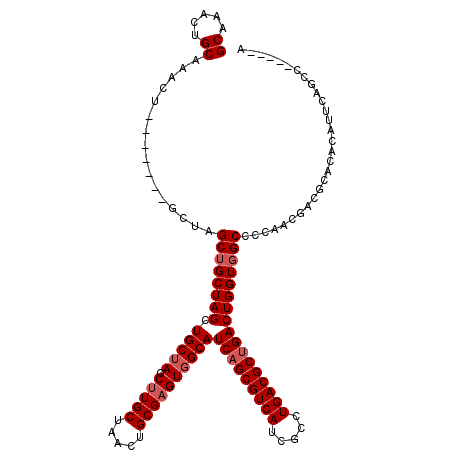
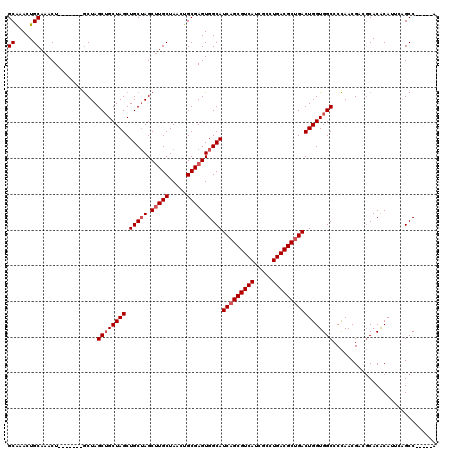
| Location | 1,405,048 – 1,405,159 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.91 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -34.39 |
| Energy contribution | -35.83 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1405048 111 - 20766785 U-----GGCUGAAUGUGUG---CGUUAGGGCCACCAGUCAGCGUCAGGCGAUGACGCUGAUGCCACUCGCAGUUAGCA-GCUUGCAGCUAGCUGCUAGUAGUUUGCAGUUUGCAGUUCGC (-----((((.((((....---))))..)))))...((((((((((.....))))))))))((.(((.(((((((((.-.......))))))))).))).))((((.....))))..... ( -45.80) >DroSec_CAF1 30878 104 - 1 U-----GGC----UGCGUGCGUCGUUGGGGCCACCAGUCAGCGUCAAGCGAUGACGCUGAUGCCACUCGCAGUUAGCAAGCUAGCAGCUAGCAGCUAGC-------AGUUUGCAGUUUGC (-----(((----((((.((((((((((.....)))).)(((((((.....))))))))))))....))))))))(((((((.(((((.....))).))-------)))))))....... ( -45.80) >DroSim_CAF1 35887 113 - 1 UUGGCUGGCUGAAUGUGUGCGUCGUUGGGGCCACCAGUCCGCGUCAGGCGAUGACGCUGAUGCCACUCGCAGUUAGCAAGCUAGCAGCUAGCAGCUAGC-------AGUUUGCAGUUUGC ...((((((((...(((.(((((....((((.....))))((((((.....)))))).))))))))...))))))))..((((((........))))))-------.............. ( -48.70) >consensus U_____GGCUGAAUGUGUGCGUCGUUGGGGCCACCAGUCAGCGUCAGGCGAUGACGCUGAUGCCACUCGCAGUUAGCAAGCUAGCAGCUAGCAGCUAGC_______AGUUUGCAGUUUGC ......(((....((.(((.(((.....))))))))((((((((((.....)))))))))))))....((((...(((((((....(((((...))))).......)))))))...)))) (-34.39 = -35.83 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:49 2006