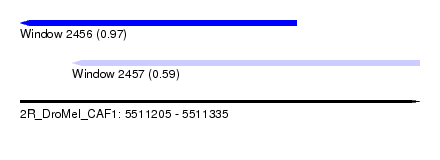
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,511,205 – 5,511,335 |
| Length | 130 |
| Max. P | 0.965746 |

| Location | 5,511,205 – 5,511,295 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.67 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
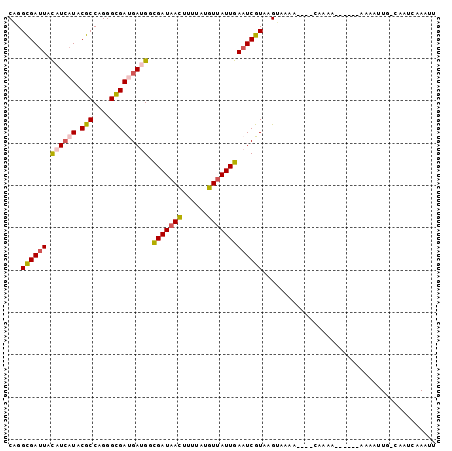
>2R_DroMel_CAF1 5511205 90 - 20766785 CGGGUGACUUUAUCAUCCGCCAGGGUGACGACGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA----CAAAA------CAAAUUU-CAAUCAAACU .(..(((......(((((....)))))((((...(((((((......)))))))..))))........----.....------.......-...)))..). ( -19.40) >DroVir_CAF1 12481 75 - 1 CGGGCGAUUACAUCAUACGCCAGGGCGAUGAUGGCGAUAAUUUUUAUGUUAUUGAAUCGUAAG-------------------------UG-CCAGCAAAUU ..((((.(((((((((.(((....))))))))..(((((((......)))))))....)))).-------------------------))-))........ ( -22.90) >DroGri_CAF1 11860 96 - 1 CAGGCGAUUUCAUCAUACGCCAGGGCGAUGAUGGUGAUAACUUUUAUGUUAUUGAAUCGUAAGUGCGC----CAAAAUGCUAGAAUAAUG-CUCUUAAAUU ..((((((((((((((.(((....)))))))))..((((((......)))))))))))((....))))----).....((.........)-)......... ( -24.20) >DroWil_CAF1 13751 94 - 1 CAGGCGAUUAUAUAAUCCGCCAGGGCGAUGAUGGUGAUAAUUUUUAUGUCAUUGAAUCGUAAGUAGACUACCCAAAA------AAAAUUG-UAAUGAAAUU ..(((((((....))).)))).(((((((...(((((((.......)))))))..))))..((....)).)))....------.......-.......... ( -17.50) >DroMoj_CAF1 12113 96 - 1 CGGGCGAUUUCAUCAUACGCCAGGGUGAUGAUGGCGAUAAUUUUUAUGUUAUUGAAUCGUGAGUACAG----CCAACGAGAAAUAAAAUG-CCAACAAAUU ..((((...(((((((.(((....))))))))))(((((((......)))))))..((((..(.....----)..)))).........))-))........ ( -23.10) >DroAna_CAF1 13555 91 - 1 CAGGCGAUUAUAUCAUUCGCCAGGGCGACGACGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA----CAAAA------CAAAUUUCCAAUCAAACU ..(((((.........))))).(((..((((...(((((((......)))))))..))))..((....----....)------).....)))......... ( -19.90) >consensus CAGGCGAUUACAUCAUACGCCAGGGCGAUGAUGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA____CAAAA______AAAAUUG_CAAUCAAAUU ...((((((.((((((.(((....))))))))).(((((((......)))))))))))))......................................... (-16.13 = -16.22 + 0.09)

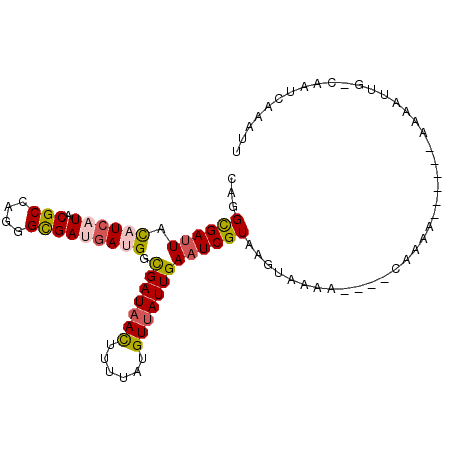
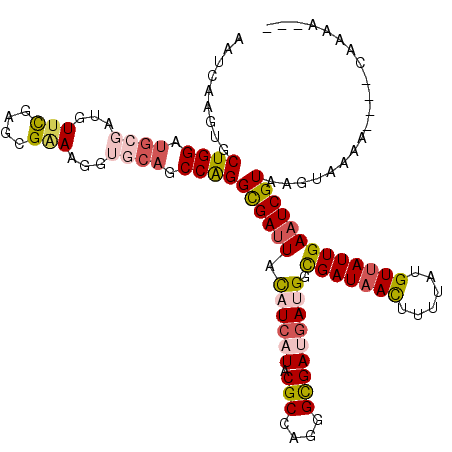
| Location | 5,511,222 – 5,511,335 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5511222 113 - 20766785 AACCAAGUCCUGGAUGCCAUGUUCGAGCGAAAGGUUCAGCCGGGUGACUUUAUCAUCCGCCAGGGUGACGACGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA----CAAAA--- .......((((((.......(((.((.(....).)).)))(((((((.....)))))))))))))..((((...(((((((......)))))))..))))........----.....--- ( -32.20) >DroVir_CAF1 12493 103 - 1 AAUCAAGUGCUGGACGCGAUGUUCGAGCGGAAGGUGCUGCCGGGCGAUUACAUCAUACGCCAGGGCGAUGAUGGCGAUAAUUUUUAUGUUAUUGAAUCGUAAG----------------- .....((..((...(((.........)))...))..)).....((((((.((((((.(((....))))))))).(((((((......)))))))))))))...----------------- ( -33.00) >DroGri_CAF1 11880 116 - 1 AAUCAAGUGCUGGAUGCGAUGUUCGAGCGGAAGGUGGAACCAGGCGAUUUCAUCAUACGCCAGGGCGAUGAUGGUGAUAACUUUUAUGUUAUUGAAUCGUAAGUGCGC----CAAAAUGC ..........(((.((((..((((.(.(....).).))))...(((((((((((((.(((....)))))))))..((((((......)))))))))))))...)))))----))...... ( -36.60) >DroWil_CAF1 13768 117 - 1 AAUCAAGUGCUGGAUGCCAUGUUUGAGCGUAAAGUGCAGCCAGGCGAUUAUAUAAUCCGCCAGGGCGAUGAUGGUGAUAAUUUUUAUGUCAUUGAAUCGUAAGUAGACUACCCAAAA--- ......(..((..((((((....)).))))..))..).(((.(((((((....))).))))..))).(((((.((((((.......))))))...))))).................--- ( -29.70) >DroMoj_CAF1 12133 116 - 1 AAUCAAGUGCUGGAUGCGAUGUUCGAGCGAAAGGUGCAGCCGGGCGAUUUCAUCAUACGCCAGGGUGAUGAUGGCGAUAAUUUUUAUGUUAUUGAAUCGUGAGUACAG----CCAACGAG ......(((((((.((((...(((....)))...)))).))..(((((((((((((.(((....))))))))))(((((((......))))))))))))).)))))..----........ ( -34.70) >DroAna_CAF1 13573 113 - 1 AACCAGGUCCUGGAUGCGAUGUUCGAGCGAAAGGUUCAGCCAGGCGAUUAUAUCAUUCGCCAGGGCGACGACGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA----CAAAA--- ..((((...)))).(((((((((.((((.....)))).(((.(((((.........)))))..)))))).....(((((((......))))))).)))))).......----.....--- ( -32.60) >consensus AAUCAAGUGCUGGAUGCGAUGUUCGAGCGAAAGGUGCAGCCAGGCGAUUACAUCAUACGCCAGGGCGAUGAUGGCGAUAACUUUUAUGUUAUUGAAUCGUAAGUAAAA____CAAAA___ .........((((.((((...(((....)))...)))).))))((((((.((((((.(((....))))))))).(((((((......))))))))))))).................... (-24.12 = -24.82 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:51 2006