
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,500,497 – 5,500,648 |
| Length | 151 |
| Max. P | 0.999776 |

| Location | 5,500,497 – 5,500,608 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.40 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
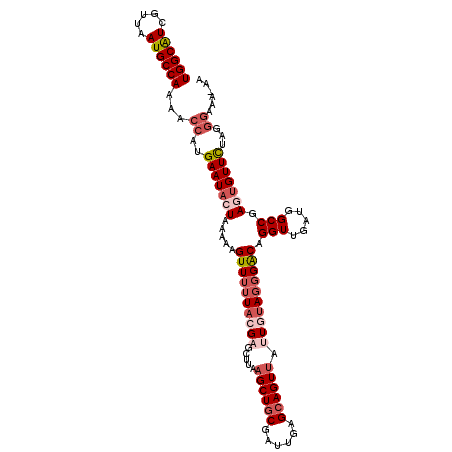
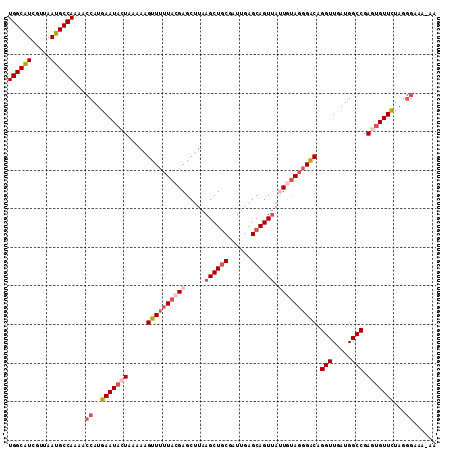
>2R_DroMel_CAF1 5500497 111 + 20766785 AUAAAACUGAAAGUAAAGCUAAAUAAAACGCUGCCAUAAUUGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCAUAAGCUGCGAUUGAGCAGUUAU ....(((((..(((..((((......((((.(((((....))))).)))).((((......(.((((.(((....))).)))).).)))).))))...)))...))))).. ( -24.50) >DroSec_CAF1 25188 111 + 1 AUAAAACUGAAAGUAAAGCUGAAUAAAACGCUGCCAUAAUUGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUAUACGAGCUUAAGCUCCGAUUGAGCAGUUAU ....((((.......(((((......((((.(((((....))))).))))............((((((........))))))...)))))..((((.....)))))))).. ( -24.00) >DroSim_CAF1 2329 111 + 1 AUAAAACUGAAAGUAAAGCUGAAUAAAACGCUGCCAUAAUUGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUAU ...............(((((......((((.(((((....))))).)))).............((((.(((....))).))))..))))).((((((......)))))).. ( -25.50) >DroEre_CAF1 2217 111 + 1 AUAAAACUGAAAGUAAAGCUAAAUAAAACGCUGCCAUAAUUGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCAAUUGGGCAGUAAC ...............(((((......((((.(((((....))))).)))).............((((.(((....))).))))..)))))..(((((......)))))... ( -24.10) >DroYak_CAF1 1883 111 + 1 AUAAAACUGAAAGUAAAGCUGAAUAAAACGCUGCCAUAAUUGGCGUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUAC ...............(((((......((((.(((((....))))).)))).............((((.(((....))).))))..))))).((((((......)))))).. ( -24.80) >consensus AUAAAACUGAAAGUAAAGCUGAAUAAAACGCUGCCAUAAUUGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUAU ....(((((..(((..((((......((((.(((((....))))).))))........................((((((....)))))).))))...)))...))))).. (-22.94 = -23.18 + 0.24)

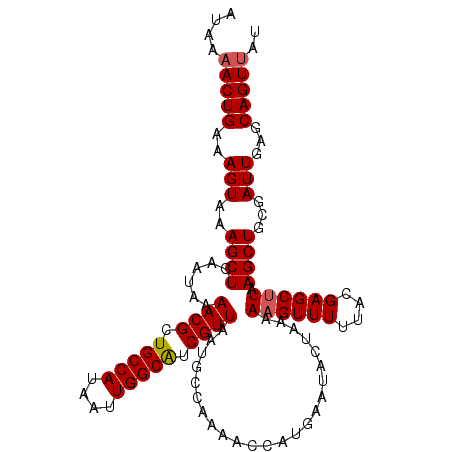
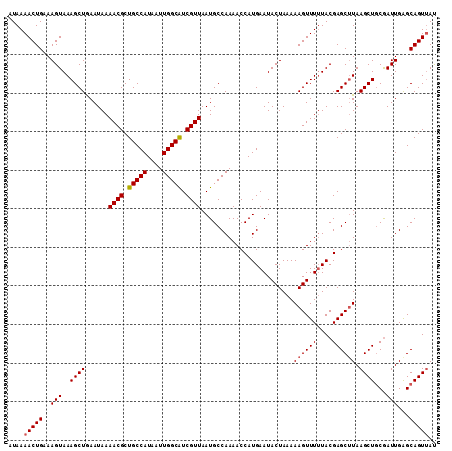
| Location | 5,500,497 – 5,500,608 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 96.40 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5500497 111 - 20766785 AUAACUGCUCAAUCGCAGCUUAUGCUCGUAAAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGAUGCCAAUUAUGGCAGCGUUUUAUUUAGCUUUACUUUCAGUUUUAU ((((((((......)))).)))).......((((((....((((.(((((((..(((((((...)))))))))))))).(((..........))).))))...)))))).. ( -23.10) >DroSec_CAF1 25188 111 - 1 AUAACUGCUCAAUCGGAGCUUAAGCUCGUAUAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGAUGCCAAUUAUGGCAGCGUUUUAUUCAGCUUUACUUUCAGUUUUAU ..(((((........((((....)))).............((((.(((((((..(((((((...)))))))))))))).(((..........))).))))..))))).... ( -22.00) >DroSim_CAF1 2329 111 - 1 AUAACUGCUCAAUCGCAGCUUAAGCUCGUAAAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGAUGCCAAUUAUGGCAGCGUUUUAUUCAGCUUUACUUUCAGUUUUAU ..(((((.......((.(((.((((.(((.((.(((....))).)).))))))).)))...((((.(((((....))))).)))).......))........))))).... ( -23.16) >DroEre_CAF1 2217 111 - 1 GUUACUGCCCAAUUGCAGCUUAAGCUCGUAAAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGAUGCCAAUUAUGGCAGCGUUUUAUUUAGCUUUACUUUCAGUUUUAU ...((((.......((.(((.((((.(((.((.(((....))).)).))))))).)))...((((.(((((....))))).)))).......))........))))..... ( -22.96) >DroYak_CAF1 1883 111 - 1 GUAACUGCUCAAUCGCAGCUUAAGCUCGUAAAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGACGCCAAUUAUGGCAGCGUUUUAUUCAGCUUUACUUUCAGUUUUAU ((((..(((.((((((.(((.((((.(((.((.(((....))).)).))))))).))).........((((....)))).)))....))).))).))))............ ( -22.80) >consensus AUAACUGCUCAAUCGCAGCUUAAGCUCGUAAAAACUUUUUAGUAUUCAUGGUUUUGGCAUUAACGAUGCCAAUUAUGGCAGCGUUUUAUUCAGCUUUACUUUCAGUUUUAU ..(((((.......((.(((.((((.(((.((.(((....))).)).))))))).)))...((((.(((((....))))).)))).......))........))))).... (-20.00 = -21.00 + 1.00)

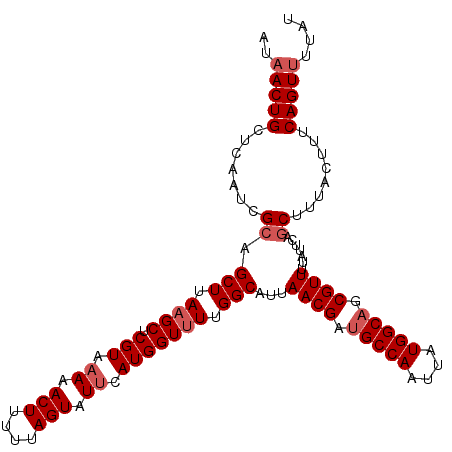
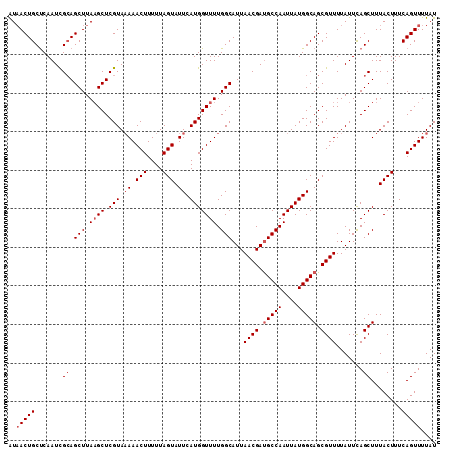
| Location | 5,500,537 – 5,500,648 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -25.64 |
| Energy contribution | -27.96 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5500537 111 + 20766785 UGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCAUAAGCUGCGAUUGAGCAGUUAUUGUAGGGACAGGUUGAUGGCCGAGUGUUCUAGGGAAAUGA ((((((.....))))))...((..(((((((.....((((((((((.....((((((......)))))).)))))))))).(((.....))).)))))))...))...... ( -36.40) >DroSec_CAF1 25228 103 + 1 UGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUAUACGAGCUUAAGCUCCGAUUGAGCAGUUAUUGUAGGGACAGGUUGAUGGCCGAGUGUUUUU--------A ((((((.....)))))).......(((((((.....(((..(((((......((((.....)))).....)))))..))).(((.....))).)))))))..--------. ( -25.80) >DroSim_CAF1 2369 111 + 1 UGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUAUUGUAGGGACAGGUUGAUGGCCGAGUGUUCUAUGGAAAAGA ((((((.....))))))...(((((((((((.....((((((((((.....((((((......)))))).)))))))))).(((.....))).))))))).))))...... ( -38.40) >DroEre_CAF1 2257 108 + 1 UGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCAAUUGGGCAGUAACUAUAGCGACAGGUUGAUGGCCGACGGUUCUAGGGC---AU (((.((((((...((((.((((.((((.(((....))).))))...(((...(((((......)))))......)))....))))..)))).)))))).)))....---.. ( -27.80) >DroYak_CAF1 1923 111 + 1 UGGCGUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUACUACAGGGGCAGGUUGAUGGCCGACUGUUCUAGGGAAAUAU ((((((.....))))))...((.((.........((((((....)))))).((((((......))))))....))(((((((.(((.....)))))))))).))....... ( -27.30) >consensus UGGCAUCGUUAAUGCCAAAACCAUGAAUACUAAAAAGUUUUUACGAGCUUAAGCUGCGAUUGAGCAGUUAUUGUAGGGACAGGUUGAUGGCCGAGUGUUCUAGGGAAA_AA ((((((.....))))))...((..(((((((.....((((((((((.....((((((......)))))).)))))))))).(((.....))).)))))))...))...... (-25.64 = -27.96 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:45 2006