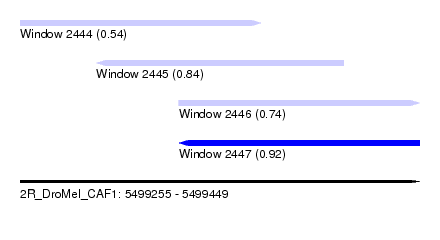
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,499,255 – 5,499,449 |
| Length | 194 |
| Max. P | 0.919912 |

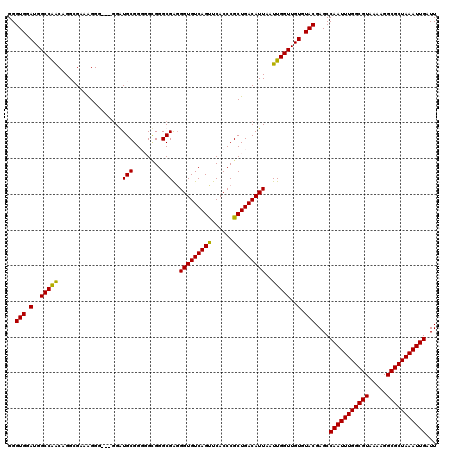
| Location | 5,499,255 – 5,499,372 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -33.31 |
| Energy contribution | -33.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5499255 117 + 20766785 GGAA---AGGGGGCGGGGGUUCAGCGGGUGAUAAAUGUGCGUGAUUUGGAAUGAAUGCGCAUCGAUUAAAACCGCAGGGCAAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACAC ....---...(..((((.((((.((((.((((..((((((((............))))))))..))))...)))).))))..(((((((.(((.......))).)))))))))))..).. ( -37.20) >DroSec_CAF1 23899 112 + 1 GG--------GGGCGGAGGUUCAGCGGGUGAUAAAUGUGCGUGAUUUGGAAUGAAUGCGCAUCGAUUAAAACCGCAGGGCAAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACAC ..--------((((....((((.((((.((((..((((((((............))))))))..))))...)))).))))...((((((.(((.......))).))))))))))...... ( -36.10) >DroEre_CAF1 921 120 + 1 GGGGUGUCGGUGGUGGGGGUUCAGCGGGUGAUAAAUGUGCGUGAUUUGGAAUGAAUGCGCAUCGAUUAAAACCGCAGGGCAAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACAC (((((((..(...(((..((((.((((.((((..((((((((............))))))))..))))...)))).))))..)))..)..)))))))....(((.....)))........ ( -40.10) >DroYak_CAF1 625 111 + 1 GGGG---------CGGGGGUUCAGCGGGUGAUAAAUGUGCGUGAUUUGGAAUGAAUGCGCAUCGAUUAAAACCGCAGGGCAAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACAC .(..---------((((.((((.((((.((((..((((((((............))))))))..))))...)))).))))..(((((((.(((.......))).)))))))))))..).. ( -37.20) >consensus GGGG______GGGCGGGGGUUCAGCGGGUGAUAAAUGUGCGUGAUUUGGAAUGAAUGCGCAUCGAUUAAAACCGCAGGGCAAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACAC ...............(((((((.((((.((((..((((((((............))))))))..))))...)))).))))..(((((((.(((.......))).))))))))))...... (-33.31 = -33.12 + -0.19)

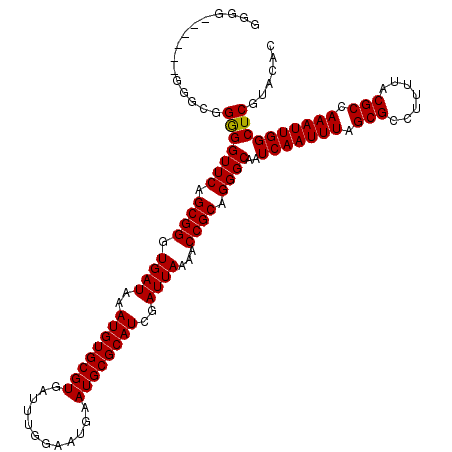
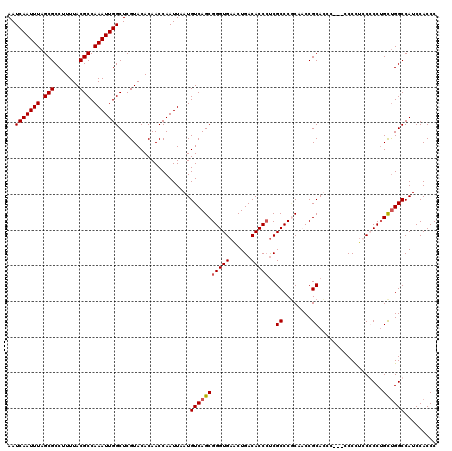
| Location | 5,499,292 – 5,499,412 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -39.29 |
| Energy contribution | -39.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5499292 120 - 20766785 UGGGCGAUGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUUGCCCUGCGGUUUUAAUCGAUGCGCAUUCAUUCCAAAUCAC ((((...((((((((((......))))))))))..((..(((((((((..(((((((((((.....)))))))))))(((((...))))).....))).))))))..)).))))...... ( -40.10) >DroSec_CAF1 23931 120 - 1 AGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUUGCCCUGCGGUUUUAAUCGAUGCGCAUUCAUUCCAAAUCAC ((((((((((((......)))))(((.((((.........))))...)))(((((((((((.....))))))))))).)))))))((((((......))).)))................ ( -39.50) >DroEre_CAF1 961 120 - 1 CGGGCGGGGGUGUCAGCUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUUGCCCUGCGGUUUUAAUCGAUGCGCAUUCAUUCCAAAUCAC .((...(((((((((((......)))))))))...((..(((((((((..(((((((((((.....)))))))))))(((((...))))).....))).))))))..))..))...)).. ( -41.60) >DroYak_CAF1 656 120 - 1 CGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUUGCCCUGCGGUUUUAAUCGAUGCGCAUUCAUUCCAAAUCAC .(((((((((((......)))))(((.((((.........))))...)))(((((((((((.....))))))))))).))))))(((((((......))).))))............... ( -39.30) >consensus CGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUUGCCCUGCGGUUUUAAUCGAUGCGCAUUCAUUCCAAAUCAC .(((....(((((((((......)))))))))...((..(((((((((..(((((((((((.....)))))))))))(((((...))))).....))).))))))..)).)))....... (-39.29 = -39.10 + -0.19)

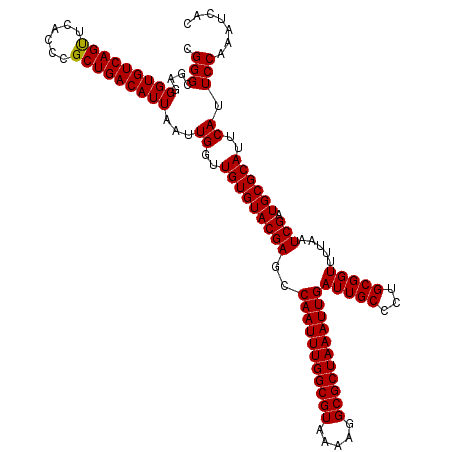
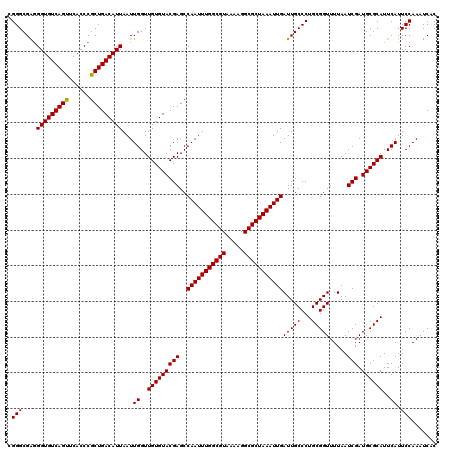
| Location | 5,499,332 – 5,499,449 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5499332 117 + 20766785 AAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACACAACCAAUUAAUGUCAGCGGGUGAACUGACACCAUCGCCCACCACCGCAUCC---CCCUUUCCCCUGUUGGCCAUCCACCC ..(((((((.(((.......))).)))))))....................(((((((((.(((..((.....))((........))....---....))).)))))))))......... ( -22.10) >DroSec_CAF1 23971 117 + 1 AAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACACAACCAAUUAAUGUCAGCGGGUGAACUGACACCCUCGCCCUCCACCGCAUCC---CCCUCUCCCCUGUUGGCCAUCCACCC ..(((((((.(((.......))).)))))))....................(((((((((((......)))))..((........))....---...........))))))......... ( -23.50) >DroEre_CAF1 1001 120 + 1 AAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACACAACCAAUUAAUGUCAGCGGGUGAGCUGACACCCCCGCCCGCACCCGCACCCGCACCCCUCCGCCUGCUGGCCAUCCACCC ..(((((((.(((.......))).)))))))....................(((((((((((.((.(.(......).).))....((....)).......)))))))))))......... ( -29.40) >DroYak_CAF1 696 115 + 1 AAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACACAACCAAUUAAUGUCAGCGGGUGAACUGACACCCUCGCCCGCACCCGCACC-----CCCUCCGCCAGCCGGCCAUCCACCC ..........(((........(((.....))).......................((((((((..........))))))))...)))...-----......(((....)))......... ( -26.50) >consensus AAUCAAUUUAGCGCCUUUUACGCCAAAUUGGCUCGUACACAACCAAUUAAUGUCAGCGGGUGAACUGACACCCUCGCCCGCAACCGCACCC___CCCCUCCCCCUGCUGGCCAUCCACCC ..(((((((.(((.......))).)))))))....................(((((((((((......)))))..((........))..................))))))......... (-22.02 = -22.28 + 0.25)

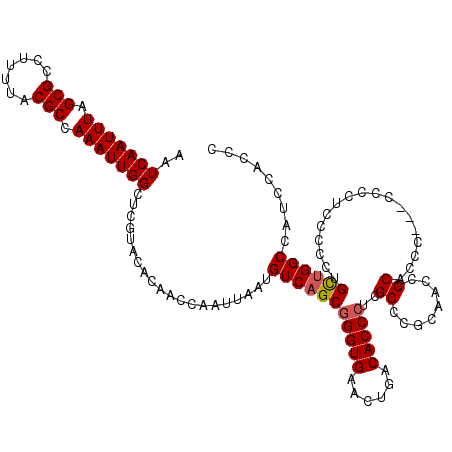
| Location | 5,499,332 – 5,499,449 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -38.72 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5499332 117 - 20766785 GGGUGGAUGGCCAACAGGGGAAAGGG---GGAUGCGGUGGUGGGCGAUGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUU ..(((.(..((((((........)..---.((((((((((((((((((...))).))))).)))))).))))..)))))..).)))....(((((((((((.....)))))))))))... ( -40.30) >DroSec_CAF1 23971 117 - 1 GGGUGGAUGGCCAACAGGGGAGAGGG---GGAUGCGGUGGAGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUU ..(((.(..(((((............---...(((........)))..(((((((((......)))))))))..)))))..).)))....(((((((((((.....)))))))))))... ( -38.00) >DroEre_CAF1 1001 120 - 1 GGGUGGAUGGCCAGCAGGCGGAGGGGUGCGGGUGCGGGUGCGGGCGGGGGUGUCAGCUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUU ..(((.(..(((((((..((........))..))).((((..(((((((((....)).).))))))..))))...))))..).)))....(((((((((((.....)))))))))))... ( -44.40) >DroYak_CAF1 696 115 - 1 GGGUGGAUGGCCGGCUGGCGGAGGG-----GGUGCGGGUGCGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUU ..(((.(..(((((((.(((..(..-----....)...))).)))...(((((((((......)))))))))...))))..).)))....(((((((((((.....)))))))))))... ( -42.30) >consensus GGGUGGAUGGCCAACAGGCGAAAGGG___GGAUGCGGGGGCGGGCGAGGGUGUCAGUUCACCCGCUGACAUUAAUUGGUUGUGUACGAGCCAAUUUGGCGUAAAAGGCGCUAAAUUGAUU ..(((.(..(((((..................(((........)))..(((((((((......)))))))))..)))))..).)))....(((((((((((.....)))))))))))... (-38.72 = -38.10 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:41 2006