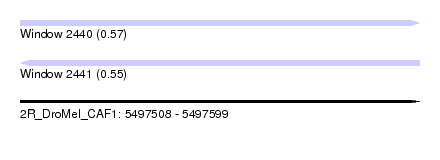
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,497,508 – 5,497,599 |
| Length | 91 |
| Max. P | 0.566902 |

| Location | 5,497,508 – 5,497,599 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5497508 91 + 20766785 GGAUGCCCACUUGAGGAGUUGUCCUUUAAU----GGGUGAGGCGCGGAAGUGCAGGACUUCCUGGUGUCCUUGAACGACCGGAGCUGAUCUAAAU (((((((((...(((((....)))))...)----))))..(((.(((..((..(((((........)))))...))..)))..))).)))).... ( -31.50) >DroSim_CAF1 16252 91 + 1 GGAUGCCCACUUGAGGAGUUGUCCUUUAAU----GGGCGAGGCGCGGAAGUGCAGGACUUCCUGGUGUCCUUGAGCGACCGGGGCAGAUCUUAAU ...((((((...(((((....)))))...)----)))))..((((....))))(((((((((((((((......)).))))))).)).))))... ( -36.70) >DroEre_CAF1 16221 95 + 1 GGAUGCCCACUUGAGGAGUUGUCCUUCCUUGGGAGGGAGGGACGCGGAAGUGCGGGACUUCCUGGUGUCCUUGAGCGAUCGGAGCAGCUCUAAGU .....(((.((..(((((......)))))..)).)))((((((((((((((.....))))))..))))))))((((..........))))..... ( -38.90) >DroYak_CAF1 17194 91 + 1 GGAUGCCCACUUGAGGAGUGGUCCUUUAAC----GGGAGACGCGCGGAAGUGCAGGACUUCCUGGUGUCCUUGACGGACCGGAGCAGCUCUAGAU ((((((((((((...))))(((((.(((..----((((.((((((....))))(((....))).)).))))))).))))))).)))..))).... ( -35.60) >consensus GGAUGCCCACUUGAGGAGUUGUCCUUUAAU____GGGAGAGGCGCGGAAGUGCAGGACUUCCUGGUGUCCUUGAGCGACCGGAGCAGAUCUAAAU (((((((.....(((((....)))))........(((((..((((....))))....))))).)))))))..........(((.....))).... (-24.46 = -25.03 + 0.56)

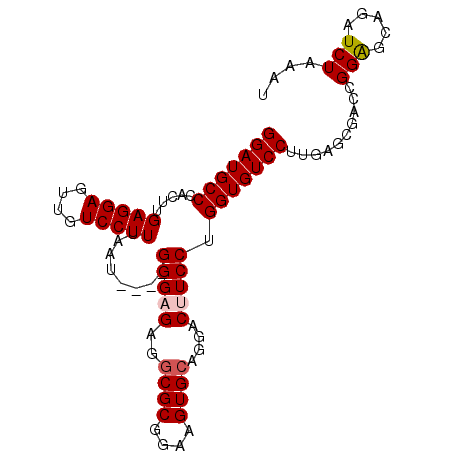
| Location | 5,497,508 – 5,497,599 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5497508 91 - 20766785 AUUUAGAUCAGCUCCGGUCGUUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCACCC----AUUAAAGGACAACUCCUCAAGUGGGCAUCC .....((...((...(((.(((....)))))).((((((.....))))))))...)).(((----(((..((((....))))..))))))..... ( -27.40) >DroSim_CAF1 16252 91 - 1 AUUAAGAUCUGCCCCGGUCGCUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCGCCC----AUUAAAGGACAACUCCUCAAGUGGGCAUCC ...............((.(((....((...)).((((((.....)))))))))))..((((----(((..((((....))))..))))))).... ( -30.10) >DroEre_CAF1 16221 95 - 1 ACUUAGAGCUGCUCCGAUCGCUCAAGGACACCAGGAAGUCCCGCACUUCCGCGUCCCUCCCUCCCAAGGAAGGACAACUCCUCAAGUGGGCAUCC .........(((((.....(((..((((.....((((((.....))))))..((((.(((.......))).))))...))))..))))))))... ( -27.70) >DroYak_CAF1 17194 91 - 1 AUCUAGAGCUGCUCCGGUCCGUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCGUCUCCC----GUUAAAGGACCACUCCUCAAGUGGGCAUCC .....(((.(((....((((.....))))....((((((.....))))))..))).)))((----......)).(((((.....)))))...... ( -27.20) >consensus AUUUAGAGCUGCUCCGGUCGCUCAAGGACACCAGGAAGUCCUGCACUUCCGCGCCUCUCCC____AUUAAAGGACAACUCCUCAAGUGGGCAUCC ..........(((((((.(((....((...)).((((((.....)))))))))))...............((((....))))...).)))).... (-17.91 = -17.98 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:35 2006