
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,490,727 – 5,490,817 |
| Length | 90 |
| Max. P | 0.543770 |

| Location | 5,490,727 – 5,490,817 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
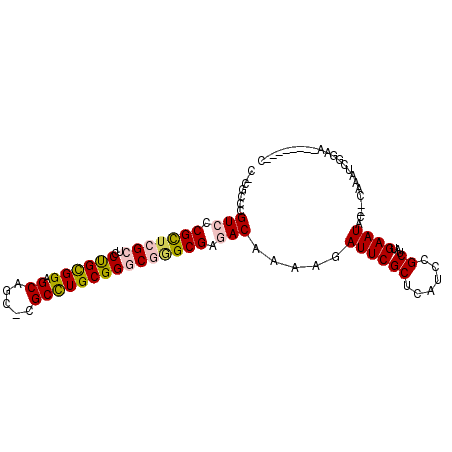
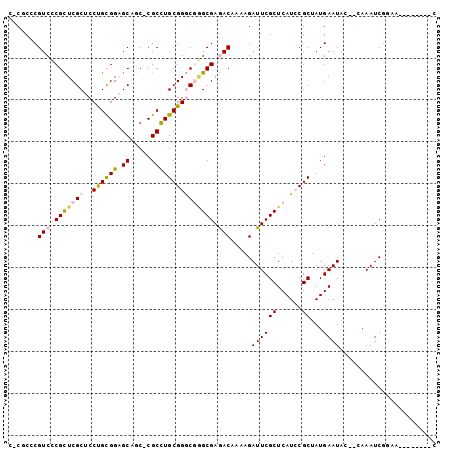
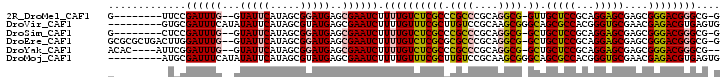
>2R_DroMel_CAF1 5490727 90 + 20766785 C-CGCCCGUCCCGCUCGCUCCUGCGGAGCAAC-CGCCUGCGGGCGGGCGAGACAAAAGAUUCGCUCAUCCGCUAUGAAUAC--CAAAUCGGAA--------C (-(((((((..((...(((((...)))))...-))...))))))))(((((.(....).)))))...((((...((.....--))...)))).--------. ( -33.60) >DroVir_CAF1 12406 93 + 1 CACUCACGUCUCGUUCGCACCCGUGGCGCUGCCCGCUUGCGGACAAGCGAAACAAAAGAUUCGCUCAUACGCUAUGAAUAUAUGAAAUCGCAC--------- ............(((((((..((.(((...)))))..)))))))..((((........((((((......))...))))........))))..--------- ( -23.59) >DroSim_CAF1 11416 90 + 1 C-CGCCCGUCCCGCUCGCUCCUGCGGAGCAGC-CGCCUGCGGGCGGGCGAGACAAAAGAUUCGCUCAUCCGCUAUGAAUAC--CAAAUCGGAG--------C (-(((((((.(.(((.(((((...))))))))-.)...))))))))(((((.(....).)))))...((((...((.....--))...)))).--------. ( -36.20) >DroEre_CAF1 9400 98 + 1 C-CGCCCGUCCCGCUCGCUCCUGCGGAGCAGC-CGCCUGCGGGCGCGCGAGACAAAAGAUUCGCUCAUCCGCUAUGAAUAC--CAAAUCCAAGUCAGCGCGC .-.((((((.(.(((.(((((...))))))))-.)...))))))((((..(((.........((......))..((.....--)).......))).)))).. ( -34.00) >DroYak_CAF1 9558 93 + 1 --CGCCCGUCCCGCUCGCUCCUGCGGAGCAGC-CGCCUGCGGGCGGGCGAGACAAAAGAUUCGCUCAUCCGCUAUGAAUAC--CAAAUCCGAAU----GUGU --((((((.(((((..((..((((...)))).-.))..))))))))))).......(.((((((......))..((.....--)).....))))----.).. ( -33.60) >DroMoj_CAF1 18008 93 + 1 CACUCACGUCUCGUUCGCACCCGUGGCGCUGCCCGCUUGCGGACAAGCGAAACAAAAGAUUCGCUCAUACGCUAUGAAUAUAUGAAAUCGCAU--------- ............(((((((..((.(((...)))))..)))))))..((((........((((((......))...))))........))))..--------- ( -23.59) >consensus C_CGCCCGUCCCGCUCGCUCCUGCGGAGCAGC_CGCCUGCGGGCGGGCGAGACAAAAGAUUCGCUCAUCCGCUAUGAAUAC__CAAAUCGGAA________C .......(((.(((((((..((((((.((.....))))))))))))))).))).....((((((......))...))))....................... (-22.82 = -22.93 + 0.11)

| Location | 5,490,727 – 5,490,817 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.78 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
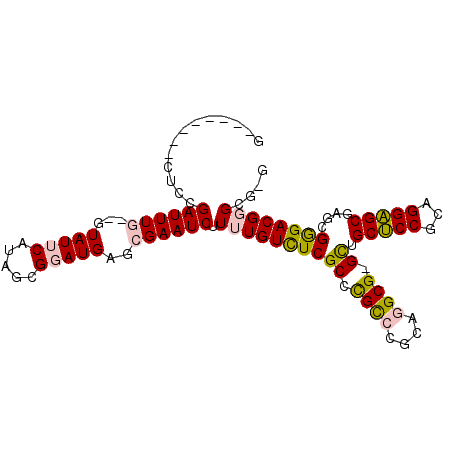
>2R_DroMel_CAF1 5490727 90 - 20766785 G--------UUCCGAUUUG--GUAUUCAUAGCGGAUGAGCGAAUCUUUUGUCUCGCCCGCCCGCAGGCG-GUUGCUCCGCAGGAGCGAGCGGGACGGGCG-G (--------(((.((((((--.(((((.....)))))..))))))....((((((((((((....))))-)(((((((...)))))))))))))))))).-. ( -40.00) >DroVir_CAF1 12406 93 - 1 ---------GUGCGAUUUCAUAUAUUCAUAGCGUAUGAGCGAAUCUUUUGUUUCGCUUGUCCGCAAGCGGGCAGCGCCACGGGUGCGAACGAGACGUGAGUG ---------.............(((((((..(((..(((((((........)))))))(((((....))))).(((((...)))))..)))....))))))) ( -29.60) >DroSim_CAF1 11416 90 - 1 G--------CUCCGAUUUG--GUAUUCAUAGCGGAUGAGCGAAUCUUUUGUCUCGCCCGCCCGCAGGCG-GCUGCUCCGCAGGAGCGAGCGGGACGGGCG-G (--------(.((((((((--.(((((.....)))))..))))))....((((((((((((....))))-).((((((...)))))).))))))))))).-. ( -41.00) >DroEre_CAF1 9400 98 - 1 GCGCGCUGACUUGGAUUUG--GUAUUCAUAGCGGAUGAGCGAAUCUUUUGUCUCGCGCGCCCGCAGGCG-GCUGCUCCGCAGGAGCGAGCGGGACGGGCG-G (((((..(((..(((((((--.(((((.....)))))..)))))))...))).)))))(((((....(.-((((((((...))))).))).)..))))).-. ( -45.30) >DroYak_CAF1 9558 93 - 1 ACAC----AUUCGGAUUUG--GUAUUCAUAGCGGAUGAGCGAAUCUUUUGUCUCGCCCGCCCGCAGGCG-GCUGCUCCGCAGGAGCGAGCGGGACGGGCG-- ..((----(...(((((((--.(((((.....)))))..)))))))..)))..(((((((((((..((.-.((((...))))..))..))))).))))))-- ( -40.80) >DroMoj_CAF1 18008 93 - 1 ---------AUGCGAUUUCAUAUAUUCAUAGCGUAUGAGCGAAUCUUUUGUUUCGCUUGUCCGCAAGCGGGCAGCGCCACGGGUGCGAACGAGACGUGAGUG ---------.............(((((((..(((..(((((((........)))))))(((((....))))).(((((...)))))..)))....))))))) ( -29.60) >consensus G________CUCCGAUUUG__GUAUUCAUAGCGGAUGAGCGAAUCUUUUGUCUCGCCCGCCCGCAGGCG_GCUGCUCCGCAGGAGCGAGCGGGACGGGCG_G .............((((((...(((((.....)))))..)))))).((((((((((.((((....)))).)).(((((...)))))....)))))))).... (-25.92 = -25.78 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:30 2006