
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,484,169 – 5,484,274 |
| Length | 105 |
| Max. P | 0.530920 |

| Location | 5,484,169 – 5,484,274 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5484169 105 + 20766785 AAAAGCCGCAAACUUCAACCUGAUAAAUGCCCACAAUCAGGAAGACGGAGGAGUCUUUCUGUUGGCGGGCACCAACUUCAACAAGGGGUUCGUUAUUAA---C-UGACU ..............(((...((((((.(((((.(((.(((((((((......))))))))))))..)))))(.((((((.....)))))).))))))).---.-))).. ( -30.20) >DroVir_CAF1 5085 107 + 1 AAAUGCCGGCAAUUUCAAUUUGAUCAAUGCCCACAAUCUGGAGGACGAUGGCGUCUUUUUGCUAGCUGGCACCAAUUUCAAUCGCGGGUGAGUUUUGAUCCCAC-AUA- ...(((((((....................(((.....)))((((((....)))))).......)))))))............(.(((...........))).)-...- ( -24.00) >DroPse_CAF1 4529 107 + 1 GAAUGCGGGUAAUUUCAACCUGAUAAAUGCCCAUAAUCUGGAGGACAAUGGCGUGUUCCUUUUGGCAGGCACUAACUACAAUCGCGGGUGAGUC-UUAAUGCU-UCAAU (((.(((......((((.((((.....((((..(((...((((.((......)).))))..)))...)))).............))))))))..-....))))-))... ( -22.57) >DroSec_CAF1 2870 105 + 1 AAAGGCCGCAAACUUCAACCUGAUAAAUGCCCACAAUCAGGAAGACGGAGGAGUCUUUCUGCUGGCGGGCACCAACUUCAACAAGGGGUUCGUUAUUAA---C-UGACU ..............(((...((((((.(((((.(...(((((((((......)))))))))...).)))))(.((((((.....)))))).))))))).---.-))).. ( -28.90) >DroSim_CAF1 2866 105 + 1 AAAGGCCGCAAACUUCAACCUGAUAAAUGCCCACAAUCAGGAAGACGGAGGAGUCUUUCUGCUGGCGGGCACCAACUUCAACAAGGGGUUCGUUAUUAA---C-UGACU ..............(((...((((((.(((((.(...(((((((((......)))))))))...).)))))(.((((((.....)))))).))))))).---.-))).. ( -28.90) >DroYak_CAF1 2740 105 + 1 GAAAGCGGCAAACUUCAACUUGAUAAAUGCCCACAACCAGGAAGACGGAGGAGUCUUUCUGCUGGCGGGCACGAACUUUAAUAAGGGGUUAGUUGUUAA---C-UGCUU ..((((((..((((.(..((((.(((((((((.(...(((((((((......)))))))))...).))))).....)))).))))..)..)))).....---)-))))) ( -34.60) >consensus AAAAGCCGCAAACUUCAACCUGAUAAAUGCCCACAAUCAGGAAGACGGAGGAGUCUUUCUGCUGGCGGGCACCAACUUCAACAAGGGGUUAGUUAUUAA___C_UGACU ...((((.(.....((.....))....(((((.(...(((((((((......)))))))))...).))))).............).))))................... (-21.32 = -21.02 + -0.30)

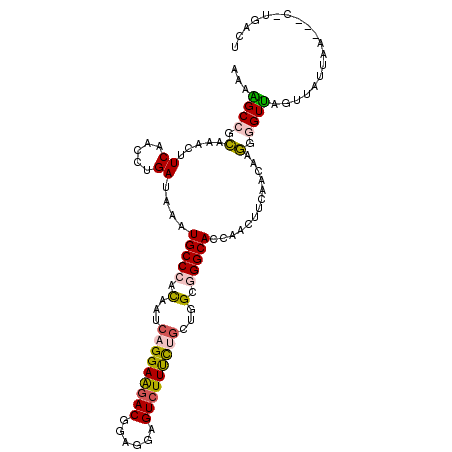
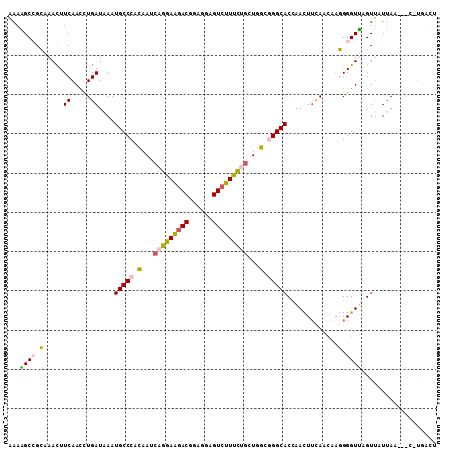
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:26 2006