| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
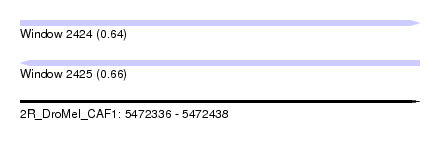
| Location | 5,472,336 – 5,472,438 |
| Length | 102 |
| Max. P | 0.656075 |

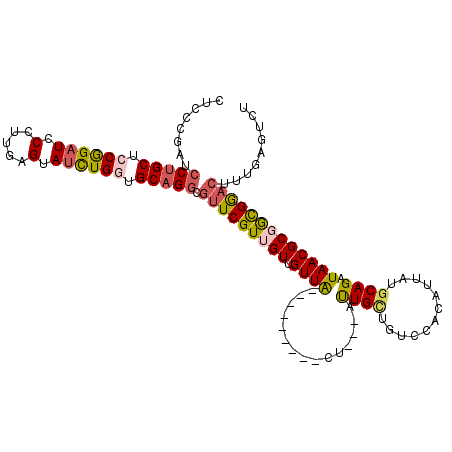
| Location | 5,472,336 – 5,472,438 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -18.36 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5472336 102 + 20766785 AGACUCAAAGUCCACAGCGUUAUCUGCAUAAUGUGGACAGCAAU---AG---------CAACAACAACGAACGCCUGCACCAGAUACUCAAGGGAUCCGGAGCAGGAUCGGGAG ...(((...((((((((((.....)))....))))))).((...---.)---------)........(((...(((((.((.(((.((....))))).)).))))).))).))) ( -32.50) >DroSec_CAF1 1 100 + 1 --ACUCAAAGUCCAUCGCGUUAUCUGCAUAAUGUGGACAGCAAU---AA---------UAACAACAACGAACGCCUGCACCAGAUACUCAAGGGAUCCGGAGCAGGAUCGGGAA --........(((.(((((((((....)))))))))........---..---------.........(((...(((((.((.(((.((....))))).)).))))).)))))). ( -27.90) >DroSim_CAF1 3 102 + 1 AGACUCAAAGUCCAUCGCGUUAUCUGCAUAAUGUGGACAGCAAU---AA---------UAACAACAACGAACGCCUGCACCAGAUACUCAAGGGAUCCGGAGCAGGAUCGGGAG ...(((...((((...(((((((....)))))))))))......---..---------.........(((...(((((.((.(((.((....))))).)).))))).))).))) ( -29.30) >DroEre_CAF1 2 102 + 1 AGACUCAAAGUUCGCAGCGUUAUCUGCAUAAUGUGGACAACAGC---AG---------UAACAACAACGAACGCCUGCAGCAAAUUCUCAAAGGAUCUGGAGCAGGAUCGGGUG ..((((...(((((..(((((.((..(.....)..)).))).))---.(---------(....))..))))).(((((..((.((((.....)))).))..)))))...)))). ( -28.80) >DroYak_CAF1 3 102 + 1 AGACUCAAAGUCCGCCGCGUUAUCUGCAGAAUGCGGACAACAAC---AG---------UAACAACAACGAACGCCUGCACCAGAUUCUCAAAGGAUCCGGGGCAGGAUCGGGUG .(((.....)))((((..(((.(((((.....))))).)))...---..---------.........(((...(((((.((.(((((.....))))).)).))))).))))))) ( -35.30) >DroPer_CAF1 306 108 + 1 AGAGUCAAAGUUCGCCGCGUUAUCUGCAAAACAUUGACAUCAGCAACAAUGCCAGCAUCAACAACAACGAGAGACUGCAGCAGAUCCUCAAAGGAGCCGGAGCCGGCU------ .(((((((.(((....(((.....)))..))).))))).)).(((....)))................(((.(((((...))).))))).....(((((....)))))------ ( -22.90) >consensus AGACUCAAAGUCCACCGCGUUAUCUGCAUAAUGUGGACAGCAAC___AA_________UAACAACAACGAACGCCUGCACCAGAUACUCAAAGGAUCCGGAGCAGGAUCGGGAG .........((((((.(((.....))).....)))))).............................(((...(((((.((.(((.((....))))).)).))))).))).... (-18.36 = -19.25 + 0.89)

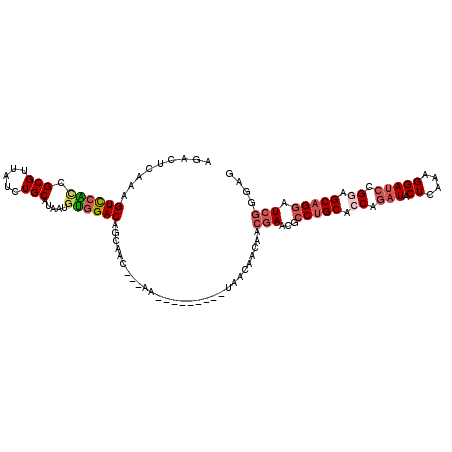
| Location | 5,472,336 – 5,472,438 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5472336 102 - 20766785 CUCCCGAUCCUGCUCCGGAUCCCUUGAGUAUCUGGUGCAGGCGUUCGUUGUUGUUG---------CU---AUUGCUGUCCACAUUAUGCAGAUAACGCUGUGGACUUUGAGUCU (((.(((.(((((.((((((.(.....).)))))).)))))...)))........(---------(.---...)).(((((((....((.......)))))))))...)))... ( -31.10) >DroSec_CAF1 1 100 - 1 UUCCCGAUCCUGCUCCGGAUCCCUUGAGUAUCUGGUGCAGGCGUUCGUUGUUGUUA---------UU---AUUGCUGUCCACAUUAUGCAGAUAACGCGAUGGACUUUGAGU-- ........(((((.((((((.(.....).)))))).))))).(((((((((.((((---------((---..(((............)))))))))))))))))).......-- ( -32.20) >DroSim_CAF1 3 102 - 1 CUCCCGAUCCUGCUCCGGAUCCCUUGAGUAUCUGGUGCAGGCGUUCGUUGUUGUUA---------UU---AUUGCUGUCCACAUUAUGCAGAUAACGCGAUGGACUUUGAGUCU (((.....(((((.((((((.(.....).)))))).))))).(((((((((.((((---------((---..(((............))))))))))))))))))...)))... ( -33.50) >DroEre_CAF1 2 102 - 1 CACCCGAUCCUGCUCCAGAUCCUUUGAGAAUUUGCUGCAGGCGUUCGUUGUUGUUA---------CU---GCUGUUGUCCACAUUAUGCAGAUAACGCUGCGAACUUUGAGUCU .....((((((((..(((((.(.....).)))))..))))).((((((.((.((((---------((---((...((....))....)))).)))))).)))))).....))). ( -29.50) >DroYak_CAF1 3 102 - 1 CACCCGAUCCUGCCCCGGAUCCUUUGAGAAUCUGGUGCAGGCGUUCGUUGUUGUUA---------CU---GUUGUUGUCCGCAUUCUGCAGAUAACGCGGCGGACUUUGAGUCU .....((((((((.((((((.(.....).)))))).))))).(((((((((.((((---------((---((...((....))....)))).))))))))))))).....))). ( -35.70) >DroPer_CAF1 306 108 - 1 ------AGCCGGCUCCGGCUCCUUUGAGGAUCUGCUGCAGUCUCUCGUUGUUGUUGAUGCUGGCAUUGUUGCUGAUGUCAAUGUUUUGCAGAUAACGCGGCGAACUUUGACUCU ------.((((((..((((..(..(((((..(((...)))..)))))..)..))))..)))))).........((.(((((.(((((((.......)))..)))).))))))). ( -33.70) >consensus CUCCCGAUCCUGCUCCGGAUCCCUUGAGUAUCUGGUGCAGGCGUUCGUUGUUGUUA_________CU___AUUGCUGUCCACAUUAUGCAGAUAACGCGGCGGACUUUGAGUCU ........(((((.((((((.(.....).)))))).))))).(((((((((.((((...............((((............)))).)))))))))))))......... (-21.95 = -22.04 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:19 2006