| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,444,780 – 5,444,873 |
| Length | 93 |
| Max. P | 0.997352 |

| Location | 5,444,780 – 5,444,873 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
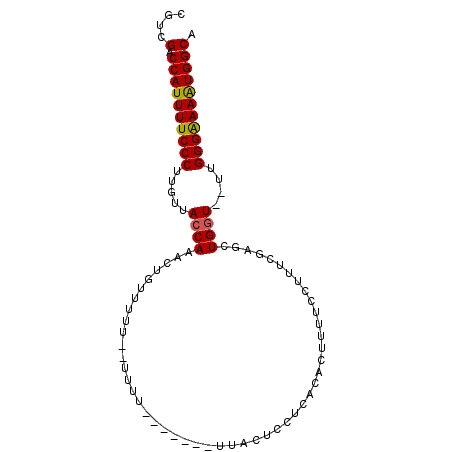
>2R_DroMel_CAF1 5444780 93 + 20766785 CGUUGACCAUUUUCCCUUGUUACCAAAUUGUUUUUUUUUUUUAUCUCUUUACUCCUCACACUUUUCCUUUCGAGCUGGU--UUGGGAAAAUGGCA ...((.((((((((((.....((((..(((........................................)))..))))--..)))))))))))) ( -19.00) >DroSec_CAF1 33232 84 + 1 CGUCGACCAUUUUCCCUUGUUACCAAACUGUUUUU--UUUU-------CAACUCCUCACACUUUUCCUUUCGAGCUGGU--UUGGGAAAAUGGCA ....(.((((((((((.....((((....(((...--....-------.)))..(((..............))).))))--..))))))))))). ( -18.84) >DroSim_CAF1 33346 84 + 1 CGUCGACCAUUUUCCCUUGUUACCAAACUGUUUUUUUUUUU-------CUACUCCUC--ACUUUUCCUUUCGAGCUGGU--UUGGGAAAAUGGCA ....(.((((((((((.....((((....((..........-------..))..(((--............))).))))--..))))))))))). ( -17.70) >DroEre_CAF1 33807 80 + 1 CGUCGACCAUUUUCCCUUGUUACCAAACGUUUU---------------UUACUUCUCACACUUUUCCUUUUCAGCUGAUUUUUGGGAAAGUGGCA ....(.((((((((((..(((....)))((...---------------..))...............................))))))))))). ( -15.00) >DroYak_CAF1 34078 78 + 1 CGUCGACCAUUUUCCCUUGUUACCAAGCAUUUU---------------UUACUUCUCACACUUUUCCUUUUGAUCUGGU--UUGGGGAAAUGGCA ....(.((((((((((.....((((..((....---------------......................))...))))--..))))))))))). ( -17.67) >consensus CGUCGACCAUUUUCCCUUGUUACCAAACUGUUUUU__UUUU_______UUACUCCUCACACUUUUCCUUUCGAGCUGGU__UUGGGAAAAUGGCA ....(.((((((((((.....((((..................................................))))....))))))))))). (-16.32 = -16.20 + -0.12)

| Location | 5,444,780 – 5,444,873 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
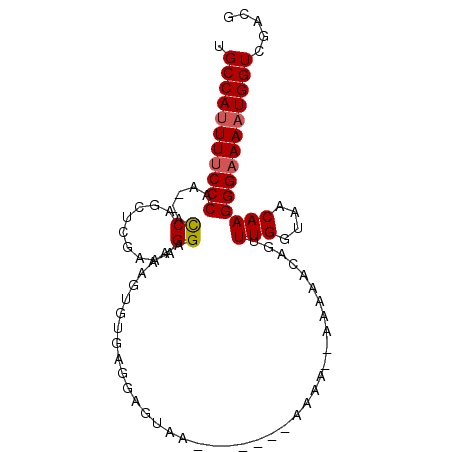
>2R_DroMel_CAF1 5444780 93 - 20766785 UGCCAUUUUCCCAA--ACCAGCUCGAAAGGAAAAGUGUGAGGAGUAAAGAGAUAAAAAAAAAAAACAAUUUGGUAACAAGGGAAAAUGGUCAACG .(((((((((((..--.((.((((....)....)))....))...........................(((....))))))))))))))..... ( -20.20) >DroSec_CAF1 33232 84 - 1 UGCCAUUUUCCCAA--ACCAGCUCGAAAGGAAAAGUGUGAGGAGUUG-------AAAA--AAAAACAGUUUGGUAACAAGGGAAAAUGGUCGACG .(((((((((((..--..((((((.................))))))-------....--.........(((....))))))))))))))..... ( -23.73) >DroSim_CAF1 33346 84 - 1 UGCCAUUUUCCCAA--ACCAGCUCGAAAGGAAAAGU--GAGGAGUAG-------AAAAAAAAAAACAGUUUGGUAACAAGGGAAAAUGGUCGACG .(((((((((((..--.((.((((....)....)))--..)).....-------...............(((....))))))))))))))..... ( -21.30) >DroEre_CAF1 33807 80 - 1 UGCCACUUUCCCAAAAAUCAGCUGAAAAGGAAAAGUGUGAGAAGUAA---------------AAAACGUUUGGUAACAAGGGAAAAUGGUCGACG .((((.((((((.....((((((..........))).))).......---------------.....(((....)))..)))))).))))..... ( -15.80) >DroYak_CAF1 34078 78 - 1 UGCCAUUUCCCCAA--ACCAGAUCAAAAGGAAAAGUGUGAGAAGUAA---------------AAAAUGCUUGGUAACAAGGGAAAAUGGUCGACG .(((((((.(((..--.((.........)).....(((...(((((.---------------....)))))....))).))).)))))))..... ( -16.40) >consensus UGCCAUUUUCCCAA__ACCAGCUCGAAAGGAAAAGUGUGAGGAGUAA_______AAAA__AAAAACAGUUUGGUAACAAGGGAAAAUGGUCGACG .(((((((((((.....((.........)).......................................(((....))))))))))))))..... (-16.22 = -16.46 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:11 2006