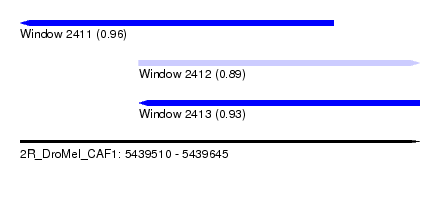
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,439,510 – 5,439,645 |
| Length | 135 |
| Max. P | 0.957509 |

| Location | 5,439,510 – 5,439,616 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5439510 106 - 20766785 GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACGAUUACGGUCAUUCGGCAUUGCAGAGCCUAGAUAACGGGACCCCACCUCCGAUCACGUUCACGUUCGGACCA (((((((......)))))))(((((........)))))..((((....(((........)))........((....)).....(((..(((....)))))))))). ( -34.30) >DroSec_CAF1 28071 99 - 1 GCCGUGGACUUAGCCACGGCGAUCGCCGAUA-ACGAUAAAGGUCAUUCGGCAUUGCAGAGCCUAGAUAACGGGUCCCCACCUCCGAUCACGUUCACGAUC------ (((((((......)))))))(((((((((..-..(((....)))..)))))......((((...(((...((((....))))...)))..))))..))))------ ( -35.60) >DroSim_CAF1 28147 100 - 1 GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACGAUAACGGUCAUUCGGCAUUGCAGAGCCUAGAUAACGGGACCCCACCUCCGAUCACGUUCACGAUC------ (((((((......)))))))(((((((((....((....)).....)))))......((((...(((..((((........)))))))..))))..))))------ ( -34.90) >DroEre_CAF1 28609 100 - 1 GCCGCGGACUUAGCCACGGCGAUCGCCGAUAAAUGAUAACGCCUAUUCGUCAAUGUAGAGCCUAGAUAACGGGACCCCACCUCCGAUCAUGUUCACGAUC------ ((((.((......)).))))(((((..((...(((((...((((((........)))).))........((((........)))))))))..)).)))))------ ( -28.20) >DroYak_CAF1 28968 100 - 1 GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACGAUAACGUUUAUUCGUCAAUGCAGAGCCUAGAUAACGGGACCCCACCUCCGAUCACGUUCACGAUC------ (((((((......)))))))(((((.(((((((((....)))))).)))........((((...(((..((((........)))))))..)))).)))))------ ( -32.80) >consensus GCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACGAUAACGGUCAUUCGGCAUUGCAGAGCCUAGAUAACGGGACCCCACCUCCGAUCACGUUCACGAUC______ (((((((......)))))))(((((((((....((....)).....)))))......((((...(((...(((......)))...)))..))))..))))...... (-28.84 = -29.68 + 0.84)

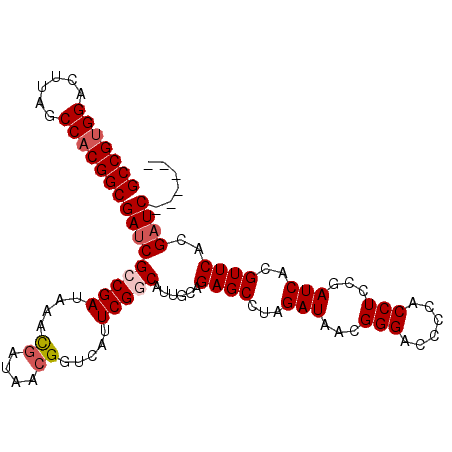

| Location | 5,439,550 – 5,439,645 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5439550 95 + 20766785 UCUAGGCUCUGCAAUGCCGAAUGACCGU-----AAU-------CGUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGCAAAUCUGCGGUGAUUAUCGCCUUC-GAAUG ((.((((.(((((.((((((.(((.((.-----...-------)).)))))))))...(((((((......))))))).....))))).(.....)))))..-))... ( -33.70) >DroSec_CAF1 28105 94 + 1 UCUAGGCUCUGCAAUGCCGAAUGACCUU-----UAU-------CGU-UAUCGGCGAUCGCCGUGGCUAAGUCCACGGCAAAUCUGCGGUGAUUAUCGCCAUC-GAAUG ....(((.(((((.((((((.((((...-----...-------.))-))))))))...(((((((......))))))).....))))).(.....))))...-..... ( -33.30) >DroSim_CAF1 28181 95 + 1 UCUAGGCUCUGCAAUGCCGAAUGACCGU-----UAU-------CGUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGCAAAUCUGCGGUGAUUAUCGCCAUC-GAAUG ....(((.(((((.((((((.(((.((.-----...-------)).)))))))))...(((((((......))))))).....))))).(.....))))...-..... ( -33.30) >DroEre_CAF1 28643 95 + 1 UCUAGGCUCUACAUUGACGAAUAGGCGU-----UAU-------CAUUUAUCGGCGAUCGCCGUGGCUAAGUCCGCGGCAAAUCUGCGCUGAUUAUCGCCUUC-GAAUG .................(((..(((((.-----...-------.....(((((((...(((((((......))))))).......)))))))...)))))))-).... ( -29.60) >DroYak_CAF1 29002 95 + 1 UCUAGGCUCUGCAUUGACGAAUAAACGU-----UAU-------CGUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGCAAAUCUGCGGUGAUUAUCGCCUUG-GAAUA (((((((.((((((((.(((.((((((.-----...-------))))))))).)))..(((((((......))))))).....))))).(.....)))).))-))... ( -33.40) >DroAna_CAF1 23724 97 + 1 -----------GAGGUCCGAAUUAUCGCCAAGCUAUCUCAAGCUAUUUUUCGAUAAUCGCAGUGGCCAAGUCCAAAGCAAAUCUAUCCCGAUUACGGCGAUCUGAAUU -----------............((((((.((((......)))).......(.((((((..((((....((.....))....))))..)))))))))))))....... ( -19.40) >consensus UCUAGGCUCUGCAAUGCCGAAUGACCGU_____UAU_______CGUUUAUCGGCGAUCGCCGUGGCUAAGUCCACGGCAAAUCUGCGGUGAUUAUCGCCAUC_GAAUG ...................................................((((((.(((((((......))))))).((((......))))))))))......... (-20.13 = -20.85 + 0.72)
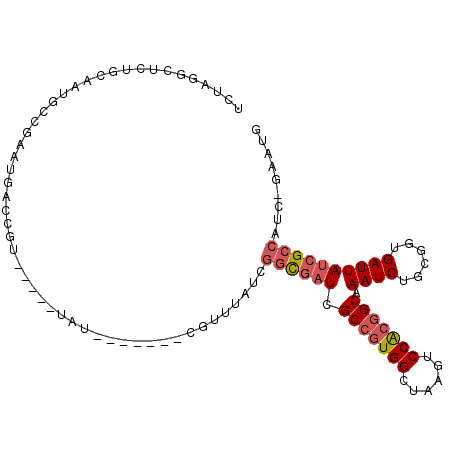
| Location | 5,439,550 – 5,439,645 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -20.33 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5439550 95 - 20766785 CAUUC-GAAGGCGAUAAUCACCGCAGAUUUGCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACG-------AUU-----ACGGUCAUUCGGCAUUGCAGAGCCUAGA .....-..(((((.....)...(((((((.(((((((......)))))))))))(((((....((-------...-----.)).....)))))..)))...))))... ( -35.00) >DroSec_CAF1 28105 94 - 1 CAUUC-GAUGGCGAUAAUCACCGCAGAUUUGCCGUGGACUUAGCCACGGCGAUCGCCGAUA-ACG-------AUA-----AAGGUCAUUCGGCAUUGCAGAGCCUAGA ...((-...((((.....)...(((((((.(((((((......)))))))))))(((((..-..(-------((.-----...)))..)))))..)))...)))..)) ( -33.70) >DroSim_CAF1 28181 95 - 1 CAUUC-GAUGGCGAUAAUCACCGCAGAUUUGCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACG-------AUA-----ACGGUCAUUCGGCAUUGCAGAGCCUAGA ...((-...((((.....)...(((((((.(((((((......)))))))))))(((((....((-------...-----.)).....)))))..)))...)))..)) ( -34.00) >DroEre_CAF1 28643 95 - 1 CAUUC-GAAGGCGAUAAUCAGCGCAGAUUUGCCGCGGACUUAGCCACGGCGAUCGCCGAUAAAUG-------AUA-----ACGCCUAUUCGUCAAUGUAGAGCCUAGA ((((.-...((((((((((......))))(((((.((......)).)))))))))))....))))-------...-----..((((((........)))).))..... ( -26.80) >DroYak_CAF1 29002 95 - 1 UAUUC-CAAGGCGAUAAUCACCGCAGAUUUGCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACG-------AUA-----ACGUUUAUUCGUCAAUGCAGAGCCUAGA .....-..(((((.....)...(((((((.(((((((......)))))))))))((.((((((((-------...-----.)))))))).))...)))...))))... ( -32.40) >DroAna_CAF1 23724 97 - 1 AAUUCAGAUCGCCGUAAUCGGGAUAGAUUUGCUUUGGACUUGGCCACUGCGAUUAUCGAAAAAUAGCUUGAGAUAGCUUGGCGAUAAUUCGGACCUC----------- .......(((((((.......(((((..((((..(((......)))..))))))))).......((((......)))))))))))............----------- ( -22.90) >consensus CAUUC_GAAGGCGAUAAUCACCGCAGAUUUGCCGUGGACUUAGCCACGGCGAUCGCCGAUAAACG_______AUA_____ACGGUCAUUCGGCAUUGCAGAGCCUAGA .........((((((((((......))))((((((((......))))))))))))))................................................... (-20.33 = -21.05 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:07 2006