| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,429,725 – 5,429,823 |
| Length | 98 |
| Max. P | 0.757415 |

| Location | 5,429,725 – 5,429,823 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
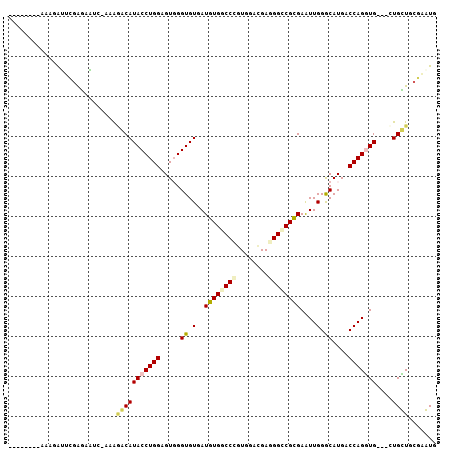
>2R_DroMel_CAF1 5429725 98 + 20766785 --------AAAGGUGCGAGAAUC-AAAUGCAUACCUGGAGUGGGUGUGAUGUGGCCCGUGGACGAGGGACGCGAGUUGGGCAUGCCCAGGUGAUGUUGCUGCGCGUG --------....((((.((.(((-(....(((((((.....))))))).((((.(((........))).))))..(((((....))))).))))....))))))... ( -32.50) >DroVir_CAF1 35199 95 + 1 -------GUAAUAGUUGAAUUU--GUGGACAUACCUGGAGUCGGUGUUAUGUGGCCGGUGGACGAGGGCCGCGAAUUGGACAUGACCAGAUG---CUGUUGCGAAUG -------((((((((.....(.--.((..((((((.......)))))..(((((((..........)))))))....)..))..)......)---)))))))..... ( -27.90) >DroGri_CAF1 33935 102 + 1 UUUUGGCGAAAUAUUUGAGAAA--UUGGACAUACCUGGUGUGGGUGUUAUAUGGCCGGUGGAUGAGGGCCGUGAAUUGGGCAUGACCAGGUG---CUGUUGCGAAUG ..((.((((....((..(....--)..))(((((((((((((...(((.(((((((..........))))))))))....))).))))))))---.)))))).)).. ( -29.90) >DroSec_CAF1 18463 98 + 1 --------AAAGGUUCGAGAAUC-AAAAGCAUACCUGGAGUGGGUGUGAUGUGGCCCGUGGACGAGGGCCGCGAGUUGGACAUGACCAGGUGAUGUUGCUGCGCGUG --------...((((....))))-...(((((((((((.(((..((...((((((((........))))))))...))..)))..)))))))....))))....... ( -33.30) >DroYak_CAF1 19406 98 + 1 --------AAAGGUGCAGGAAUC-AAAACCAUACCUGGAGUGGGUGUGAUGUGGCCCGUGGAUGAGGGUCGCGAGUUGGGCAUGACCAGGUGGUGCUGCUGCGCGUG --------....((((((....(-((...(((((((.....))))))).((((((((........))))))))..)))(((((.((...)).))))).))))))... ( -38.60) >DroMoj_CAF1 25082 95 + 1 -------A-AAUAA-AGAAUUUCGCUGGACAUACCUGGUGUCGGUGUUAUGUGACCUGUUGAGGAGGGCCGCGAAUUGGGCAUGACCAGAUG---CUGUUGCGAAUG -------.-.....-.....(((((.((.....)).((((((.(.(((((((..((..(((.((....)).)))...)))))))))).))))---))...))))).. ( -24.40) >consensus ________AAAGAUUCGAGAAUC_AAAGACAUACCUGGAGUGGGUGUGAUGUGGCCCGUGGACGAGGGCCGCGAAUUGGGCAUGACCAGGUG___CUGCUGCGAAUG ...........................(((((((((((.....((.(..((((((((........))))))))....).))....)))))))....))))....... (-20.07 = -20.23 + 0.17)


| Location | 5,429,725 – 5,429,823 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -20.11 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
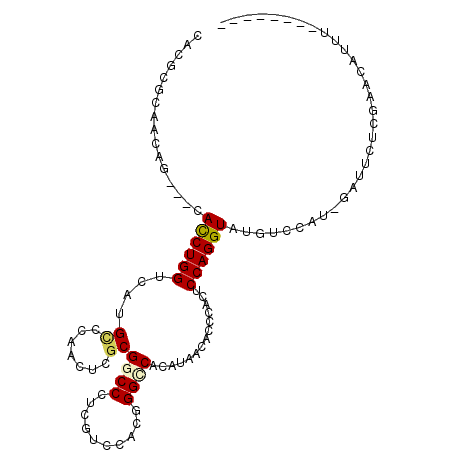
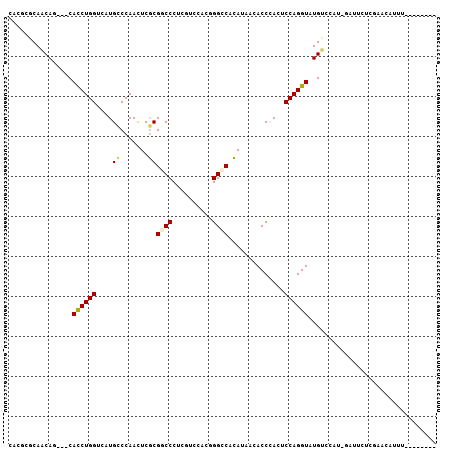
>2R_DroMel_CAF1 5429725 98 - 20766785 CACGCGCAGCAACAUCACCUGGGCAUGCCCAACUCGCGUCCCUCGUCCACGGGCCACAUCACACCCACUCCAGGUAUGCAUUU-GAUUCUCGCACCUUU-------- ...(((.((....((((..((((....))))....((((.(((.(.....(((..........)))...).))).))))...)-))).)))))......-------- ( -21.60) >DroVir_CAF1 35199 95 - 1 CAUUCGCAACAG---CAUCUGGUCAUGUCCAAUUCGCGGCCCUCGUCCACCGGCCACAUAACACCGACUCCAGGUAUGUCCAC--AAAUUCAACUAUUAC------- ........(((.---.((((((....(((......(.((((..........)))).)........))).)))))).)))....--...............------- ( -16.14) >DroGri_CAF1 33935 102 - 1 CAUUCGCAACAG---CACCUGGUCAUGCCCAAUUCACGGCCCUCAUCCACCGGCCAUAUAACACCCACACCAGGUAUGUCCAA--UUUCUCAAAUAUUUCGCCAAAA ........(((.---.(((((((..............((((..........)))).............))))))).)))....--...................... ( -16.93) >DroSec_CAF1 18463 98 - 1 CACGCGCAGCAACAUCACCUGGUCAUGUCCAACUCGCGGCCCUCGUCCACGGGCCACAUCACACCCACUCCAGGUAUGCUUUU-GAUUCUCGAACCUUU-------- ..((.(...((((((.((((((...((........(.(((((........))))).)........))..)))))))))...))-)...).)).......-------- ( -22.39) >DroYak_CAF1 19406 98 - 1 CACGCGCAGCAGCACCACCUGGUCAUGCCCAACUCGCGACCCUCAUCCACGGGCCACAUCACACCCACUCCAGGUAUGGUUUU-GAUUCCUGCACCUUU-------- .....(((((((.(((((((((....((.......))(.(((........))).)..............))))))..))).))-)....))))......-------- ( -22.40) >DroMoj_CAF1 25082 95 - 1 CAUUCGCAACAG---CAUCUGGUCAUGCCCAAUUCGCGGCCCUCCUCAACAGGUCACAUAACACCGACACCAGGUAUGUCCAGCGAAAUUCU-UUAUU-U------- ..(((((.(((.---.(((((((...((((.....).)))...........(((........)))...))))))).)))...))))).....-.....-.------- ( -21.20) >consensus CACGCGCAACAG___CACCUGGUCAUGCCCAACUCGCGGCCCUCGUCCACGGGCCACAUAACACCCACUCCAGGUAUGUCCAU_GAUUCUCGAACAUUU________ ................((((((....((.......))((((..........))))..............))))))................................ (-12.51 = -12.53 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:02 2006