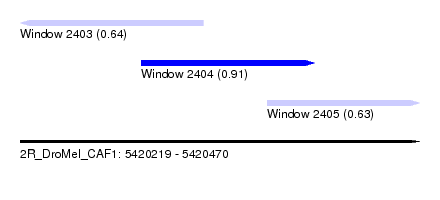
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,420,219 – 5,420,470 |
| Length | 251 |
| Max. P | 0.909612 |

| Location | 5,420,219 – 5,420,334 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.09 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5420219 115 - 20766785 UAGCCAGGUCCAUUGGGGAUGGUACAAAGGUGCGCUGCU-----GUACUUCACUUUCUUCUCUCUGAUGUGACUGAAAACACAUAAAACAUUGGGGUCACCAAACCCUUUUCGCCUGCUA ((((.((((.....(((..((((....(((((((....)-----))))))...........(((..((((...((......))....))))..)))..))))..))).....)))))))) ( -32.30) >DroSec_CAF1 9080 98 - 1 UAGCCAGGUCCAUUGGGAAUGGUGCAAAGGUG--CUGCU-----GUACUAUACUUUCUUCCCUCUGAUGUGACUG---------------UUGGGGUCACCAAACCCUUUCCUUCUGGUA ..(((((((((((((((((.((((....((((--(....-----))))).))))...)))))...)))).))).(---------------..(((((......)))))..)...))))). ( -30.90) >DroSim_CAF1 9198 98 - 1 AAGCCAGGUCCAUUGGGAAUGGUGCAAAGGUG--CUGCU-----GUACUAUACUUUCUUCCCUCUGAUGUGACUG---------------UUGGGGUCACCAAACCCUUUCCGCCUGGUA ..(((((((.....(((..(((((....((((--(....-----)))))............(((..((......)---------------)..))).)))))..))).....))))))). ( -33.30) >DroEre_CAF1 9205 100 - 1 UAGCCAGGUCCAUUCGGAGUGGUGCAAAAG----CUACUAAAAUCUACUCCACUUUCUUCUGUCUGAUGUGACUGAC--GCCAUAAGGCAUUGGGGUCACA-------------CU-GCA (((.((((.......(((((((.((....)----).........))))))).......)))).))).(((((((((.--(((....))).))..)))))))-------------..-... ( -26.54) >consensus UAGCCAGGUCCAUUGGGAAUGGUGCAAAGGUG__CUGCU_____GUACUACACUUUCUUCCCUCUGAUGUGACUG_______________UUGGGGUCACCAAACCCUUUCCGCCUGGUA ..(((((((.((..(((((((((((...(((.....))).....)))))).......)))))..))..((((((....................))))))............))))))). (-19.52 = -20.09 + 0.56)

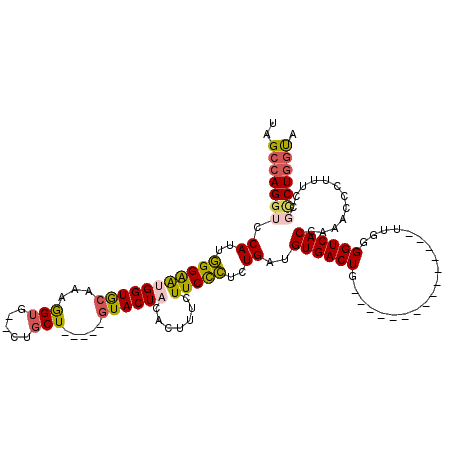
| Location | 5,420,295 – 5,420,404 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -18.13 |
| Energy contribution | -19.70 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5420295 109 + 20766785 -AGCAGCGCACCUUUGUACCAUCCCCAAUGGACCUGGCUAUUCAAUUUUGAACACUCAUGAGUGCCGCUAGGUCCUGCUACCUGGUAGCUGCAACCUGUUGCAGAUGCCA -((((..(((....)))............(((((((((..((((....))))((((....))))..)))))))))))))....((((.((((((....)))))).)))). ( -38.40) >DroSec_CAF1 9141 107 + 1 -AGCAG--CACCUUUGCACCAUUCCCAAUGGACCUGGCUAUUCAAUUUUGAAAGCUCAUGAGUGCCGCUGGGUCCUGCUACCUGCCAGCUGCAACCUGUUGCAGAUGCCA -(((((--.((((..((.((((.....))))....(((.(((((....(((....))))))))))))).)))).)))))....((...((((((....))))))..)).. ( -35.50) >DroSim_CAF1 9259 107 + 1 -AGCAG--CACCUUUGCACCAUUCCCAAUGGACCUGGCUUUUCAAUUUUGAAUGCUCAUGAGUGCCGCUGGGUCCUGCUACCUGCCAGCUGCAACCUGUUGCAGAUGCCA -(((((--.((((..((.((((.....))))....(((..((((....(((....))))))).))))).)))).)))))....((...((((((....))))))..)).. ( -33.80) >DroEre_CAF1 9269 91 + 1 UAGUAG----CUUUUGCACCACUCCGAAUGGACCUGGCUAUUCAAUUUUGAAGAC-------UGCUCCU-GGUCCUGCUACCU-------GCAACCUGUUGCACAUGCCA ..((((----(.((((........)))).(((((.(((..((((....))))...-------.)))...-))))).))))).(-------((((....)))))....... ( -23.50) >DroYak_CAF1 9375 86 + 1 ----------------CACCAUUCCUAAUGGACCUGGCUAUUCCACUUCGAAGACUCCUGACUGCCGCU-GGUCCUGCUACCU-------GCAACCUGUUGCAGAUGCCA ----------------..((((.....))))....(((..(((......))).........((((.((.-(((..(((.....-------)))))).)).))))..))). ( -21.00) >consensus _AGCAG__CACCUUUGCACCAUUCCCAAUGGACCUGGCUAUUCAAUUUUGAAGACUCAUGAGUGCCGCU_GGUCCUGCUACCUG__AGCUGCAACCUGUUGCAGAUGCCA ...............(((...........(((((((((..(((......))).(((....)))...))))))))).............((((((....)))))).))).. (-18.13 = -19.70 + 1.57)

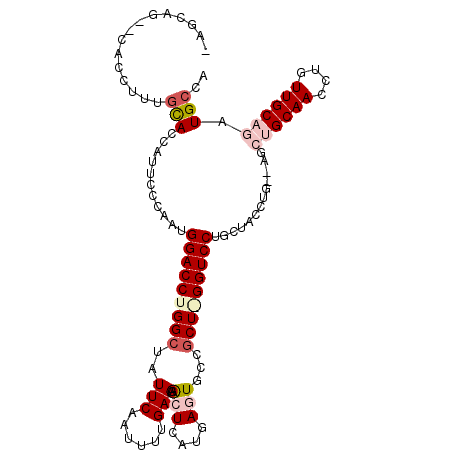
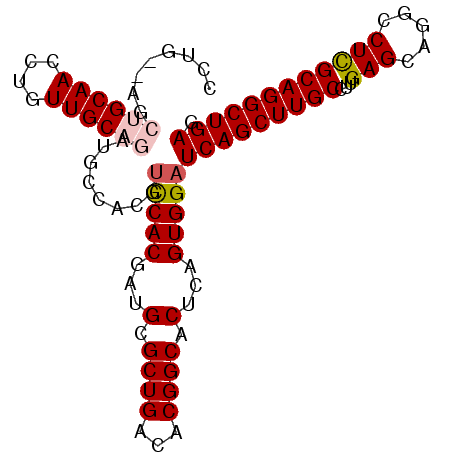
| Location | 5,420,374 – 5,420,470 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -29.64 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5420374 96 + 20766785 CCUGGUAGCUGCAACCUGUUGCAGAUGCCACGUUCACAAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUCGCAGGCUGAG ..(((((.((((((....)))))).)))))..(((((...(.((((...)))).)...)))))(((((((((...(((.....)))))))))))). ( -37.30) >DroSec_CAF1 9218 96 + 1 CCUGCCAGCUGCAACCUGUUGCAGAUGCCACGUCCACGAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUCGCAGGCUGAG (((((.((((((((....))))))..(((..((((((...(.((((...)))).)...))))))..((((.....)))).))))).)))))..... ( -37.50) >DroSim_CAF1 9336 96 + 1 CCUGCCAGCUGCAACCUGUUGCAGAUGCCACGUCCACGAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUCGCAGGCUGAG (((((.((((((((....))))))..(((..((((((...(.((((...)))).)...))))))..((((.....)))).))))).)))))..... ( -37.50) >DroEre_CAF1 9337 89 + 1 CCU-------GCAACCUGUUGCACAUGCCACGACCACGAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUUGCAGGCUGAG (((-------((((...((.((....)).))((((((...(.((((...)))).)...)))).)).((((((.....)))))).)))))))..... ( -31.60) >DroYak_CAF1 9438 89 + 1 CCU-------GCAACCUGUUGCAGAUGCCACGUCCACAAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUCGCAGGCUGAG .((-------((((....))))))........(((((...(.((((...)))).)...)))))(((((((((...(((.....)))))))))))). ( -33.70) >consensus CCUG__AGCUGCAACCUGUUGCAGAUGCCACGUCCACGAUGCGCUGACACGGCACUCAGUGGAUCAGCUUGCCUUGAGCAGGCCUCGCAGGCUGAG ........((((((....))))))........(((((...(.((((...)))).)...)))))(((((((((...(((.....)))))))))))). (-29.64 = -30.32 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:00 2006