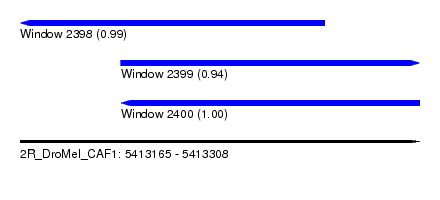
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,413,165 – 5,413,308 |
| Length | 143 |
| Max. P | 0.999948 |

| Location | 5,413,165 – 5,413,274 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -23.32 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5413165 109 - 20766785 AAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCAACAAUGUCAACUCCAUUUUAGAGAUCAUAUUUUCUG ....(..(((((((.(((..(((((((...)))))))..))))))..........))))..).....(((........)))..........((((((......)))))) ( -27.80) >DroSec_CAF1 2135 109 - 1 AAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCAACAAUGUCAACUCCAUUUUAAAGAUCAUAUUUUCUG ....(..(((((((.(((..(((((((...)))))))..))))))..........))))..).....(((........))).............(((........))). ( -25.50) >DroSim_CAF1 2222 109 - 1 AAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCAACAAUGUCAACUCCAUUUUAGAGAUCAUAUUUUCUG ....(..(((((((.(((..(((((((...)))))))..))))))..........))))..).....(((........)))..........((((((......)))))) ( -27.80) >DroEre_CAF1 2339 100 - 1 AAAAGUGCACUUGGCGGCUAUGGCCAUUACGUGGUCGCAGCC--------ACAUAAGUGUCCAAAAAGAUCUAACAAUGUCAACUCCAUUUUAAAGAUCAUAUU-UCUG ....(..(((((((.((((.(((((((...))))))).))))--------.).))))))..).....(((((...((((.......))))....))))).....-.... ( -28.90) >DroYak_CAF1 2340 100 - 1 AAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCC--------ACAUAAGUGUCCAAAAAGGCCCAACAAUGUCAACUCCAUUUUUAAGAUCAUAAU-UUUG ((((((((((((((.(((..(((((((...)))))))..)))--------.).))))))........(((........))).....)))))))...........-.... ( -26.10) >consensus AAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCAACAAUGUCAACUCCAUUUUAAAGAUCAUAUUUUCUG .((((((...((((((((..(((((((...)))))))..)))..............(.(((......))).)......)))))...))))))................. (-23.32 = -22.68 + -0.64)

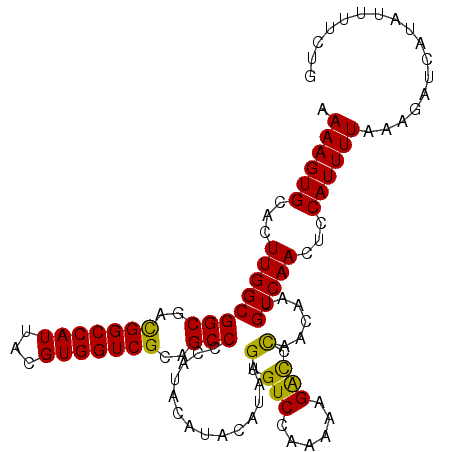
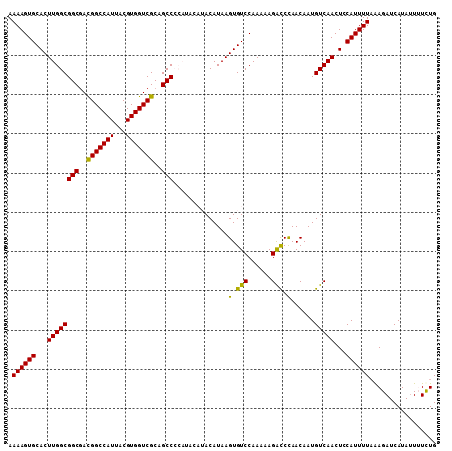
| Location | 5,413,201 – 5,413,308 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
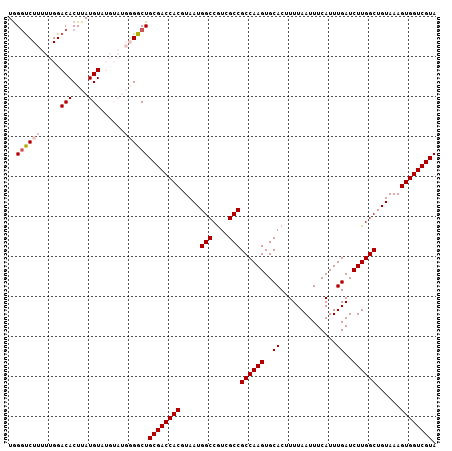
>2R_DroMel_CAF1 5413201 107 + 20766785 UGGGUCUUUUUGGACACUUAUGUAUGUAUGGGGCUGCGACCACGUAAUGGCCGUCGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ...((((....)))).((((((....))))))..(((((((((.....(((....)))((((((..((..............))..))))))......))))))))) ( -32.44) >DroSec_CAF1 2171 107 + 1 UGGGUCUUUUUGGACACUUAUGUAUGUAUGGGGCUGCGACCACGUAAUGGCCGUCGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ...((((....)))).((((((....))))))..(((((((((.....(((....)))((((((..((..............))..))))))......))))))))) ( -32.44) >DroSim_CAF1 2258 107 + 1 UGGGUCUUUUUGGACACUUAUGUAUGUAUGGGGCUGCGACCACGUAAUGGCCGUCGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ...((((....)))).((((((....))))))..(((((((((.....(((....)))((((((..((..............))..))))))......))))))))) ( -32.44) >DroEre_CAF1 2374 99 + 1 UAGAUCUUUUUGGACACUUAUGU--------GGCUGCGACCACGUAAUGGCCAUAGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ...........((.(((....))--------).))((((((((.....(((....)))((((((..((..............))..))))))......)))))))). ( -29.24) >DroYak_CAF1 2375 99 + 1 UGGGCCUUUUUGGACACUUAUGU--------GGCUGCGACCACGUAAUGGCCGUCGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ..((((.....(....)......--------))))((((((((.....(((....)))((((((..((..............))..))))))......)))))))). ( -30.84) >consensus UGGGUCUUUUUGGACACUUAUGUAUGUAUGGGGCUGCGACCACGUAAUGGCCGUCGCCGCCAAGUGCACUUUUAAUUUCAUUUGAUCUUGGCUGUAAAGUGGUCGUA ..((((((.....(((....)))......))))))((((((((.....(((....)))((((((..((..............))..))))))......)))))))). (-28.96 = -29.80 + 0.84)

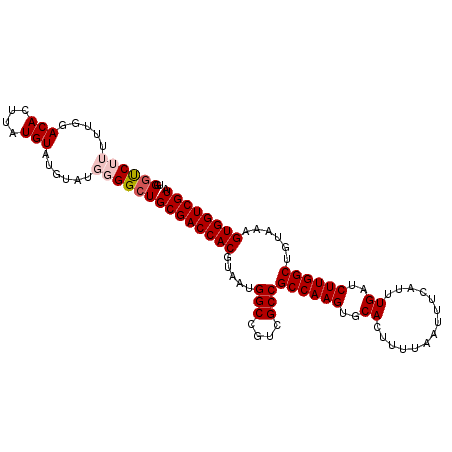
| Location | 5,413,201 – 5,413,308 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
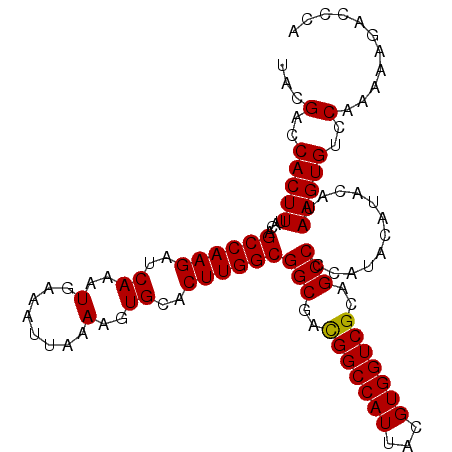
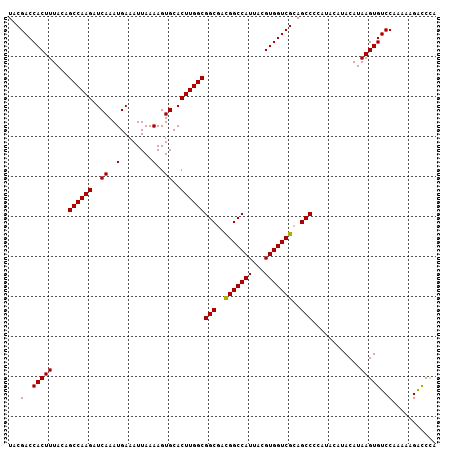
>2R_DroMel_CAF1 5413201 107 - 20766785 UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCA ...(..(((((....((((((..((..(........)..))..))))))(((..(((((((...)))))))..)))............)))))..)........... ( -27.90) >DroSec_CAF1 2171 107 - 1 UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCA ...(..(((((....((((((..((..(........)..))..))))))(((..(((((((...)))))))..)))............)))))..)........... ( -27.90) >DroSim_CAF1 2258 107 - 1 UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCA ...(..(((((....((((((..((..(........)..))..))))))(((..(((((((...)))))))..)))............)))))..)........... ( -27.90) >DroEre_CAF1 2374 99 - 1 UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCUAUGGCCAUUACGUGGUCGCAGCC--------ACAUAAGUGUCCAAAAAGAUCUA ...(..(((((....((((((..((..(........)..))..))))))((((.(((((((...))))))).))))--------....)))))..)........... ( -27.80) >DroYak_CAF1 2375 99 - 1 UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCC--------ACAUAAGUGUCCAAAAAGGCCCA ...............(((....((....))........(..(((((((.(((..(((((((...)))))))..)))--------.).))))))..).....)))... ( -29.10) >consensus UACGACCACUUUACAGCCAAGAUCAAAUGAAAUUAAAAGUGCACUUGGCGGCGACGGCCAUUACGUGGUCGCAGCCCCAUACAUACAUAAGUGUCCAAAAAGACCCA ...(..(((((....((((((..((..(........)..))..))))))(((..(((((((...)))))))..)))............)))))..)........... (-27.06 = -26.90 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:55 2006