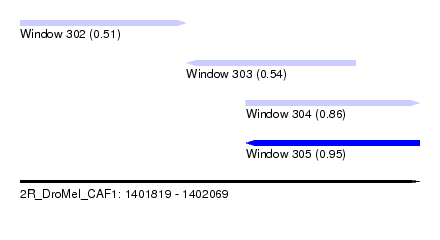
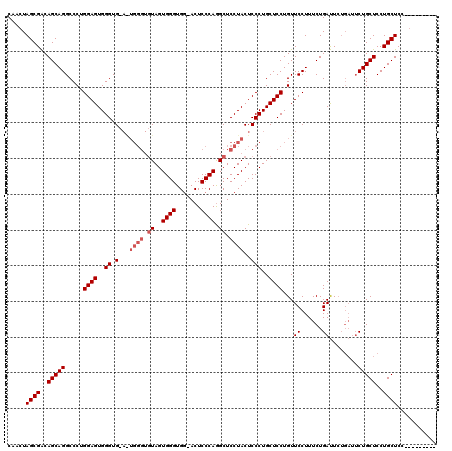
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,401,819 – 1,402,069 |
| Length | 250 |
| Max. P | 0.948541 |

| Location | 1,401,819 – 1,401,923 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -27.83 |
| Energy contribution | -29.50 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1401819 104 + 20766785 CUA-UAGCGGCAGCAGGCCCUGGAGUGGGUGGAAUGGGUGUAGUGGGUGAAACUCCCAAGCUCCUACUCCCUGCUCCUGUUCCUUUCUGACU------CUGCUCCUGCUCC--------- ...-....((.((((((...((((((.((.((((((((.((((.(((((...............))))).)))).))))))))...)).)))------)))..))))))))--------- ( -40.76) >DroSec_CAF1 27683 110 + 1 CAACUAGCGACAGCAGGCCCUGGAGUGGGUG---------UAGUGGGUGG-ACUCCCAGGCUCCUACUCCCUGCUCCUGUUCCUUUCUGAUUCUGAUUCUGCUCCUGCUCCUGCUCCUGC ......(((..((((((....((((..((.(---------(((.((((((-(((....)).))).)))).)))).))..)))).................((....)).))))))..))) ( -36.50) >DroSim_CAF1 32634 110 + 1 CAACUAGCGACAGCAGGCCCUGGAGUGGGUGUAGUGGGUGUAGUGGGUGG-ACUCCCAGGCUCCUACUCCCUGCUCCUGUUCCUUUCUGAUUCUGAUUCUGCUCCUGCUCC--------- .....((((..((((((....((((..((.(.((((((.((..((((...-...)))).)).)))))))))..))))........((.......)).))))))..))))..--------- ( -40.50) >consensus CAACUAGCGACAGCAGGCCCUGGAGUGGGUG_A_UGGGUGUAGUGGGUGG_ACUCCCAGGCUCCUACUCCCUGCUCCUGUUCCUUUCUGAUUCUGAUUCUGCUCCUGCUCC_________ .....((((..(((((.....((((..((.(...((((.((..((((.......)))).)).))))..)))..))))...((......))........)))))..))))........... (-27.83 = -29.50 + 1.67)

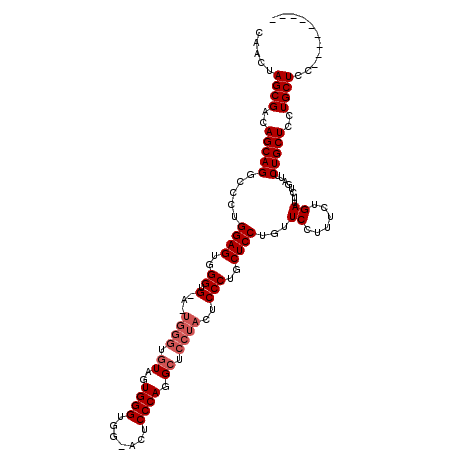
| Location | 1,401,923 – 1,402,029 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1401923 106 - 20766785 AC----------ACACA-CAGCGACAUUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCUUUGUCGCCACUUGGCAGUCGAUGCGGAUGCAGGAACA--- ..----------.(((.-((.((((...)))).))((((..((((........))))..))))....)))(((.(((((.(((.((((....))).).))).)))))))).......--- ( -28.40) >DroSec_CAF1 27793 111 - 1 ACA--------CACACA-CAGCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCUUUGUCGCCACUUGGCUGUCGAUGCGGAUGCAGGAACAGGG ...--------...((.-((.((((...)))).))))...............(((((........((.((...((((((.(((.((((....))).).))).)))))).)).))))))). ( -29.30) >DroEre_CAF1 26656 108 - 1 UCACACACAC---------ACCGACACUGGCGGUGGCAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCUUUGUCGCCACUUGGCUGUGGAUGCGGACGCAGGAGCG--- ..((.(((.(---------((((.......)))))......((((........))))..........)))))..(((((((..(.(((....))).)..)).)))))(((....)))--- ( -29.40) >DroYak_CAF1 28448 117 - 1 ACACAUACACACACACACCACCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCUUUGUUGCCACUUGGCUGUGGAUGCGGAUGCAGGAACA--- .....(((.(((....(((((((((...)))))))))....((((........))))..........))))))((((((((..(.(((....))).)..)).)))))).........--- ( -37.60) >consensus ACA_________ACACA_CACCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCUUUGUCGCCACUUGGCUGUCGAUGCGGAUGCAGGAACA___ ..................(((((((...)))))))((....((((........))))........((.((...((((((((..(.(((....))).)..)).)))))).)).)))).... (-23.35 = -24.23 + 0.87)

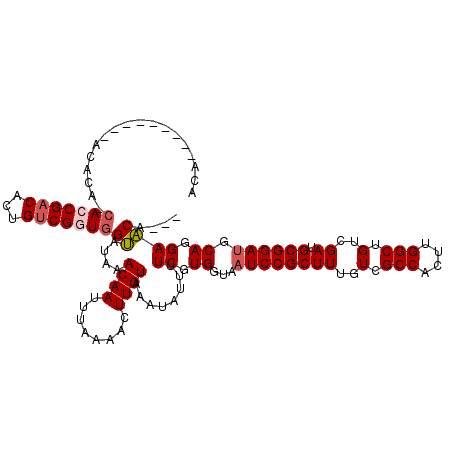
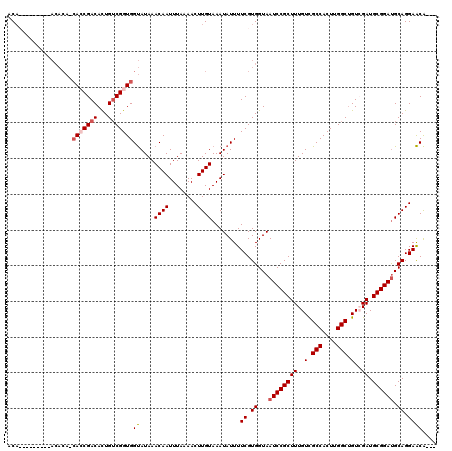
| Location | 1,401,960 – 1,402,069 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -23.33 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1401960 109 + 20766785 AGCGGAUUACCACGAAAAUAUUUACAAGUUUUAAAUUGUUUAUACCACCGACAAUGUCGCUG-UGUGU----------GUGUGUUAUCUGGCAUAUUAUUAUUUAUGCCAUUGGGCAUCA .((((((.(((((..........((((........))))..((((((.((((...)))).))-.))))----------))).)).))))((((((........)))))).....)).... ( -25.50) >DroSec_CAF1 27833 111 + 1 AGCGGAUUACCACGAAAAUAUUUACAAGUUUUAAAUUGUUUAUACCACCGACAGUGUCGCUG-UGUGUG--------UGUGUGUUAUCUGGCAUAUUAUUAUUUAUGCCAUUAGGCAUCA .((((((.((((((....(((..((((........))))..))).(((..((((.....)))-)..)))--------)))).)).))))((((((........)))))).....)).... ( -26.90) >DroEre_CAF1 26693 111 + 1 AGCGGAUUACCACGAAAAUAUUUACAAGUUUUAAAUUGUUUAUGCCACCGCCAGUGUCGGU---------GUGUGUGUGAGUGUUAUCCGGCGUAUUAUUAUUUAUGCCAUUGGGCAUCG .(((((((((((((....(((..((((........))))..))).(((((((......)))---------).))))))).)))..))))((((((........)))))).....)).... ( -30.00) >DroYak_CAF1 28485 120 + 1 AGCGGAUUACCACGAAAAUAUUUACAAGUUUUAAAUUGUUUAUACCACCGACAGUGUCGGUGGUGUGUGUGUGUAUGUGUGUGUUAUCCGGCAUAUUAUUAUUUAUGCAAUUGGGCAUCG .((((((.((((((...((((.((((..............((((((((((((...)))))))))))))))).)))).)))).)).)))).(((((........)))))......)).... ( -38.93) >consensus AGCGGAUUACCACGAAAAUAUUUACAAGUUUUAAAUUGUUUAUACCACCGACAGUGUCGCUG_UGUGU_________UGUGUGUUAUCCGGCAUAUUAUUAUUUAUGCCAUUGGGCAUCA .((((((.(((((.....(((..((((........))))..))).(((((((...)))))))................))).)).))))((((((........)))))).....)).... (-23.33 = -24.20 + 0.87)

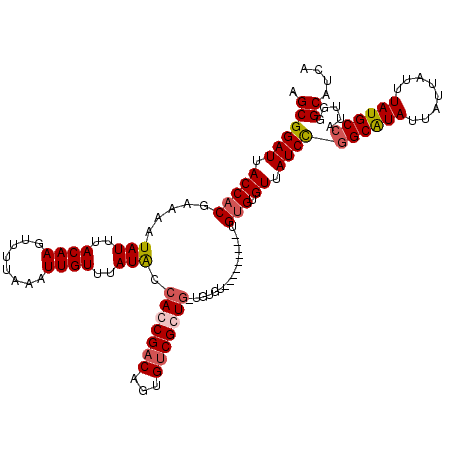
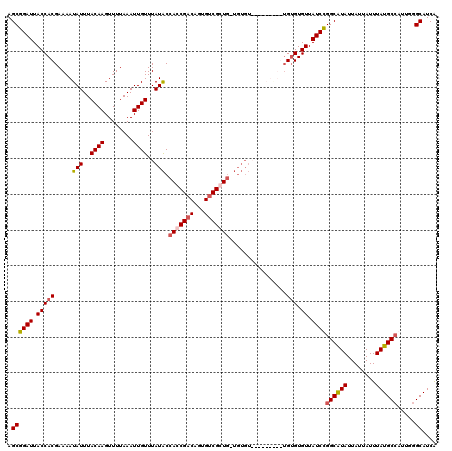
| Location | 1,401,960 – 1,402,069 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1401960 109 - 20766785 UGAUGCCCAAUGGCAUAAAUAAUAAUAUGCCAGAUAACACAC----------ACACA-CAGCGACAUUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCU ....((....(((((((........)))))))(((.((.(((----------.....-((.((((...)))).))((((..((((........))))..))))....))))).))).)). ( -23.00) >DroSec_CAF1 27833 111 - 1 UGAUGCCUAAUGGCAUAAAUAAUAAUAUGCCAGAUAACACACA--------CACACA-CAGCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCU ....((....(((((((........)))))))(((.((.(((.--------......-((.((((...)))).))((((..((((........))))..))))....))))).))).)). ( -23.00) >DroEre_CAF1 26693 111 - 1 CGAUGCCCAAUGGCAUAAAUAAUAAUACGCCGGAUAACACUCACACACAC---------ACCGACACUGGCGGUGGCAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCU ..(((((....)))))............((.((((..(((.........(---------((((.......)))))......((((........))))..........)))...)))))). ( -22.90) >DroYak_CAF1 28485 120 - 1 CGAUGCCCAAUUGCAUAAAUAAUAAUAUGCCGGAUAACACACACAUACACACACACACCACCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCU ............(((((........)))))(((((.((.(((..............(((((((((...)))))))))....((((........))))..........))))).))))).. ( -30.30) >consensus CGAUGCCCAAUGGCAUAAAUAAUAAUAUGCCAGAUAACACACA_________ACACA_CACCGACACUGUCGGUGGUAUAAACAAUUUAAAACUUGUAAAUAUUUUCGUGGUAAUCCGCU ..(((((...(((((((........)))))))...........................((((((...)))))))))))..((((........))))..........((((....)))). (-19.09 = -19.90 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:17 2006