
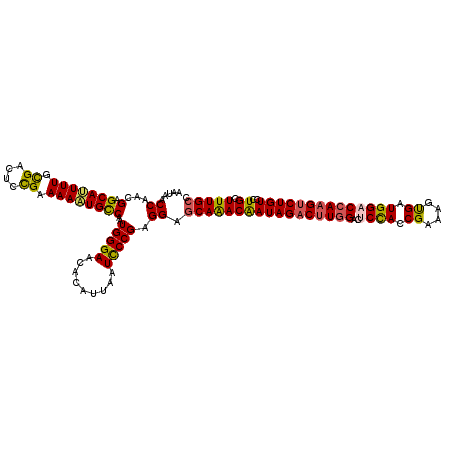
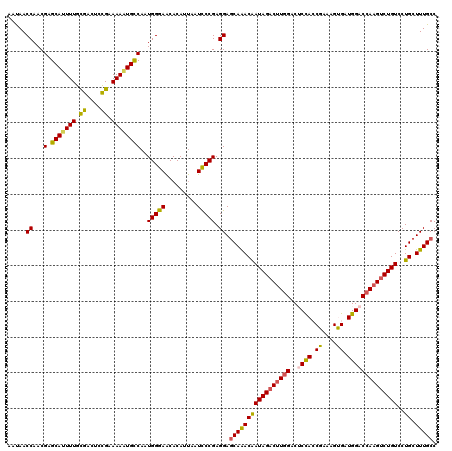
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,408,888 – 5,409,020 |
| Length | 132 |
| Max. P | 0.998121 |

| Location | 5,408,888 – 5,409,008 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -30.20 |
| Energy contribution | -29.84 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5408888 120 + 20766785 AAUAACCAACGCGCAUUUUGCGACUCCGAAAAAUGCCAAUGGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACUCUACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCC .....((...(.(((((((.((....)).))))))))..(((((.........))))).)).(((((((((((((((((..((((((....).).))))))))))))))..)).))))). ( -37.30) >DroSec_CAF1 10608 120 + 1 AAUAACCAACGAGCAUUUUGUGACUCCGAAAAAUGCCAAUGGGAACACAUUACUUCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCC .....((...(.(((((((.((....)).))))))))....((((........))))..)).(((((((((((((((((..((((((....).).))))))))))))))..)).))))). ( -36.00) >DroSim_CAF1 9105 120 + 1 AAUAACCAACGAGCAUUUUGUGACUCCGAAAAAUGCCAAUGGGAACACAUUACUUCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCC .....((...(.(((((((.((....)).))))))))....((((........))))..)).(((((((((((((((((..((((((....).).))))))))))))))..)).))))). ( -36.00) >DroEre_CAF1 10536 120 + 1 AACAACCAACGCGCAAUUUGCGACACCGAAAAUUGCCAGUGGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACACCAACGACAGCGAUGGACCAAGCCUGUCCUGCUUUGCC .....((...(.(((((((.((....)).))))))))..(((((.........))))).)).(((((((((((.(((((...(((.((....)).))).))))).))))..)).))))). ( -32.20) >DroYak_CAF1 9407 120 + 1 AAUAACCAACGGGCAAUUUGCGAAACUGAAAAAUGUCAUUGGGAGCACAUCAAUCCCGAGGAACAGACGAUAGACAUAGACGCCAACGAAAGCGAUGGACCAAGUCUGUCCUGCUUUGCC ...........(((((...(((................((((((.........))))))(((.(((((......(((...(((........))))))......))))))))))).))))) ( -29.80) >consensus AAUAACCAACGAGCAUUUUGCGACUCCGAAAAAUGCCAAUGGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCC .....((...(.(((((((.((....)).))))))))..(((((.........))))).)).(((((((((((((((((..((((.((....)).))))))))))))))..)).))))). (-30.20 = -29.84 + -0.36)

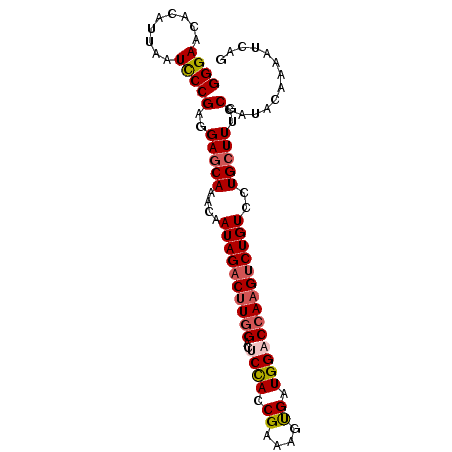
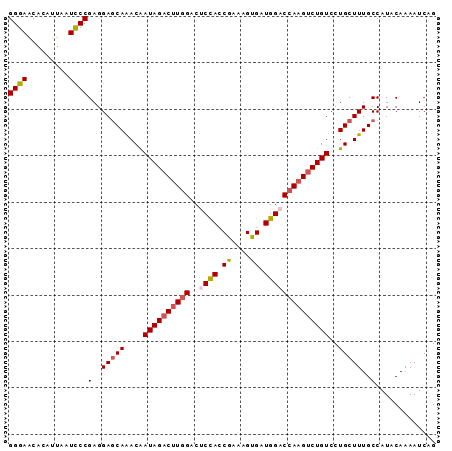
| Location | 5,408,928 – 5,409,020 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5408928 92 + 20766785 GGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACUCUACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCCAUACAAAAUCAG ((((.........))))(..(((((....((((((((((..((((((....).).))))))))))))))..)))))..)............. ( -27.70) >DroSec_CAF1 10648 92 + 1 GGGAACACAUUACUUCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCCAUACAAAAUCAG .((((........))))(..(((((....((((((((((..((((((....).).))))))))))))))..)))))..)............. ( -28.30) >DroSim_CAF1 9145 92 + 1 GGGAACACAUUACUUCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCCAUACAAAAUCGG .((((........))))(..(((((....((((((((((..((((((....).).))))))))))))))..)))))..)............. ( -28.30) >DroEre_CAF1 10576 92 + 1 GGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACACCAACGACAGCGAUGGACCAAGCCUGUCCUGCUUUGCCAUACAAAAUCAG ((((.........))))(..(((((....((((.(((((...(((.((....)).))).))))).))))..)))))..)............. ( -22.70) >DroYak_CAF1 9447 92 + 1 GGGAGCACAUCAAUCCCGAGGAACAGACGAUAGACAUAGACGCCAACGAAAGCGAUGGACCAAGUCUGUCCUGCUUUGCCAUACAGAAUCAG ((((.........)))).((((.(((((......(((...(((........))))))......))))))))).................... ( -21.20) >consensus GGGAACACAUUAAUCCCGAGGAGCAAACAAUAGACUUGGACUCCACCGAAAGUGAUGGACCAAGUCUGUCCUGCUUUGCCAUACAAAAUCAG ((((.........))))(..(((((....((((((((((..((((.((....)).))))))))))))))..)))))..)............. (-21.70 = -22.26 + 0.56)

| Location | 5,408,928 – 5,409,020 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5408928 92 - 20766785 CUGAUUUUGUAUGGCAAAGCAGGACAGACUUGGUCCAUCACUUUCGGUAGAGUCCAAGUCUAUUGUUUGCUCCUCGGGAUUAAUGUGUUCCC .((((((((...((((((.(((...((((((((.(((((......))).).).)))))))).)))))))))...)))))))).......... ( -25.60) >DroSec_CAF1 10648 92 - 1 CUGAUUUUGUAUGGCAAAGCAGGACAGACUUGGUCCAUCACUUUCGGUGGAGUCCAAGUCUAUUGUUUGCUCCUCGGAAGUAAUGUGUUCCC ....(((((...((((((.(((...((((((((((((((......))))))..)))))))).)))))))))...)))))............. ( -28.80) >DroSim_CAF1 9145 92 - 1 CCGAUUUUGUAUGGCAAAGCAGGACAGACUUGGUCCAUCACUUUCGGUGGAGUCCAAGUCUAUUGUUUGCUCCUCGGAAGUAAUGUGUUCCC ((((.((((.....))))(((((..((((((((((((((......))))))..))))))))....)))))...))))............... ( -29.10) >DroEre_CAF1 10576 92 - 1 CUGAUUUUGUAUGGCAAAGCAGGACAGGCUUGGUCCAUCGCUGUCGUUGGUGUCCAAGUCUAUUGUUUGCUCCUCGGGAUUAAUGUGUUCCC .((((((((...((((((.(((...((((((((.(((.((....)).)))...)))))))).)))))))))...)))))))).......... ( -27.90) >DroYak_CAF1 9447 92 - 1 CUGAUUCUGUAUGGCAAAGCAGGACAGACUUGGUCCAUCGCUUUCGUUGGCGUCUAUGUCUAUCGUCUGUUCCUCGGGAUUGAUGUGCUCCC ..(((((((...((...((((((..((((.(((.....((((......)))).))).))))....)))))))).)))))))........... ( -22.30) >consensus CUGAUUUUGUAUGGCAAAGCAGGACAGACUUGGUCCAUCACUUUCGGUGGAGUCCAAGUCUAUUGUUUGCUCCUCGGGAUUAAUGUGUUCCC .((((((((...((...((((((..((((((((.(((((......)))))...))))))))....)))))))).)))))))).......... (-24.48 = -24.72 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:51 2006