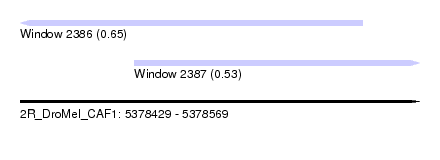
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,378,429 – 5,378,569 |
| Length | 140 |
| Max. P | 0.653231 |

| Location | 5,378,429 – 5,378,549 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.72 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
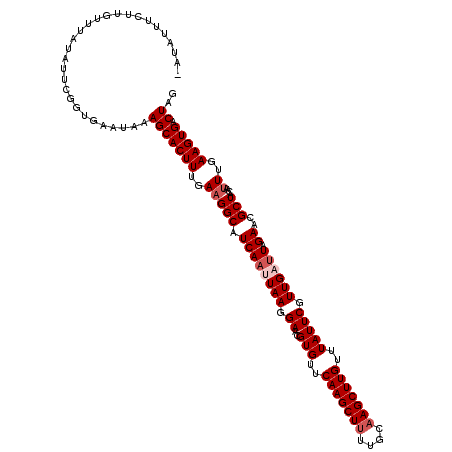
>2R_DroMel_CAF1 5378429 120 - 20766785 UGUUUUUCUUGUUUAUAUCCCGUGAAUAAAGCACUUUGAAGGCAUCAAUUAAGGAACGUGUUCAAGCUUUUGUAAGCUUGUUUAUUCGUUGAUUAGAACGCUACAAUUUGAAGUGACUAG ........((((((((.....))))))))(((((((..(((((.((((((((.((..(((..(((((((....)))))))..))))).)))))).))..)))....))..))))).)).. ( -28.80) >DroSec_CAF1 912 119 - 1 -AUAUUUCUUGUUUAUAUUCGGUGAAUAAAGCACUUUGAAGGCAUCAAUUAAGGAACGUGUUCAAGCUUUUGCAAGCUUGUUUAUUCGUUGAUUAGAACGCUACAAUUUGAAGUGACUAG -.......((((((((.....))))))))(((((((..(((((.((((((((.((..(((..(((((((....)))))))..))))).)))))).))..)))....))..))))).)).. ( -28.40) >DroSim_CAF1 1100 119 - 1 -AUAUUUCUUGUUUAUAUUCGGUGAAUAAAGCACUUUGAAGGCAUCAAUUAAGGAACGUGUUCAAGCUUUUGCAAGCUUGUUUAUUCGUUGAUUAGAACGCUACAAUUUGAAGUGACUAG -.......((((((((.....))))))))(((((((..(((((.((((((((.((..(((..(((((((....)))))))..))))).)))))).))..)))....))..))))).)).. ( -28.40) >DroEre_CAF1 1106 118 - 1 -UUAUUUCUAGCUUAUAUUCGGCGAAUAAAGCACUUUGAAGGCAUCAGUUAAGGAACGUGUUCAAGCU-UUGCAAGCUUGUUUAUUCGUUGUAUAGAACGCUACAAUUUGAAGUGACUAG -(((((((((((...(((.(((((((((((((((..(((.....)))(((....))))))).((((((-.....))))))))))))))))).)))....))))......))))))).... ( -28.00) >DroYak_CAF1 1107 114 - 1 -ACAUAU-----UUAUAUUCGGCGAAUAAAGCACUUUGAUGGCAUCAAUUAAGGAACGUGUUCAAGCUUUUGCAAGCUUGUUUAUUCGUUGUUUAGAACGCUACAAUUUGAAGUGACUAG -......-----...((..(((((((((((...(((((((.......)))))))........(((((((....))))))))))))))))))..))...((((.(.....).))))..... ( -25.60) >consensus _AUAUUUCUUGUUUAUAUUCGGUGAAUAAAGCACUUUGAAGGCAUCAAUUAAGGAACGUGUUCAAGCUUUUGCAAGCUUGUUUAUUCGUUGAUUAGAACGCUACAAUUUGAAGUGACUAG .............................(((((((..(((((.((((((((.((..(((..(((((((....)))))))..))))).)))))).))..)))....))..))))).)).. (-22.72 = -23.72 + 1.00)

| Location | 5,378,469 – 5,378,569 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -10.34 |
| Energy contribution | -10.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
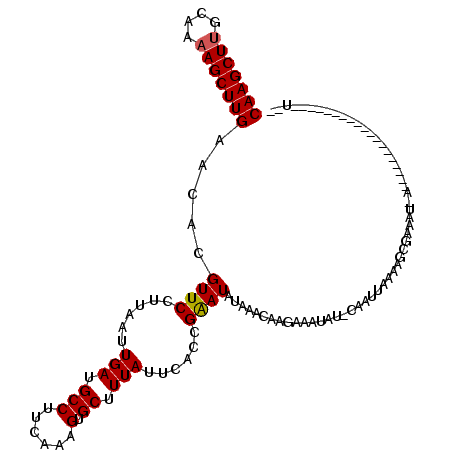
>2R_DroMel_CAF1 5378469 100 + 20766785 CAAGCUUACAAAAGCUUGAACACGUUCCUUAAUUGAUGCCUUCAAAGUGCUUUAUUCACGGGAUAUAAACAAGAAAAACACAAUUAUUA-CUAAAUUA-------------------UGU (((((((....))))))).....(((((....((((.....)))).(((.......)))))))).........................-........-------------------... ( -14.70) >DroSec_CAF1 952 96 + 1 CAAGCUUGCAAAAGCUUGAACACGUUCCUUAAUUGAUGCCUUCAAAGUGCUUUAUUCACCGAAUAUAAACAAGAAAUAU-CAAUUGAAAGCGAAAU-A---------------------- (((((((....)))))))....((((..((((((((((..(((...((....(((((...)))))...))..))).)))-))))))).))))....-.---------------------- ( -21.80) >DroSim_CAF1 1140 96 + 1 CAAGCUUGCAAAAGCUUGAACACGUUCCUUAAUUGAUGCCUUCAAAGUGCUUUAUUCACCGAAUAUAAACAAGAAAUAU-CAAUUGAAAGCGAAAU-A---------------------- (((((((....)))))))....((((..((((((((((..(((...((....(((((...)))))...))..))).)))-))))))).))))....-.---------------------- ( -21.80) >DroEre_CAF1 1146 116 + 1 CAAGCUUGCAA-AGCUUGAACACGUUCCUUAACUGAUGCCUUCAAAGUGCUUUAUUCGCCGAAUAUAAGCUAGAAAUAA-CUAUUUAAAGUAAAAA-C-ACACCCACUUCACAUUUAUCC ((((((.....-))))))................((((......(((((((((((((...))))).))))........(-((......))).....-.-......))))......)))). ( -13.30) >DroYak_CAF1 1147 113 + 1 CAAGCUUGCAAAAGCUUGAACACGUUCCUUAAUUGAUGCCAUCAAAGUGCUUUAUUCGCCGAAUAUAA-----AUAUGU-CUAUUAAAAGCGUAAA-UGGCACACACUUGAUAUUUGUCU (((((((....)))))))................((((..(((((.(((((.....(((.....(((.-----......-.))).....)))....-.)))))....)))))...)))). ( -20.50) >consensus CAAGCUUGCAAAAGCUUGAACACGUUCCUUAAUUGAUGCCUUCAAAGUGCUUUAUUCACCGAAUAUAAACAAGAAAUAU_CAAUUAAAAGCGAAAU_A___________________U__ (((((((....))))))).....((((......(((.(((......).)).)))......))))........................................................ (-10.34 = -10.38 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:42 2006