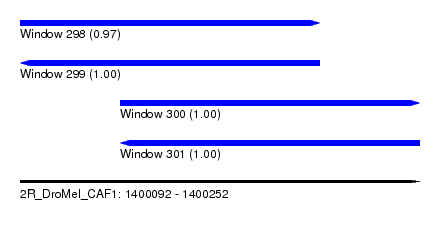
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,400,092 – 1,400,252 |
| Length | 160 |
| Max. P | 0.999924 |

| Location | 1,400,092 – 1,400,212 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1400092 120 + 20766785 ACUCACACUUCAUUUAGCCAGAUGUUGCACAUCCGAACGGCAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAU ................(((((.(((((.....((....))...((((((((.....(((((....)))))...))))))))......................)))))...))))).... ( -25.30) >DroSec_CAF1 25896 120 + 1 GCUCACACUUCAUUUAGCCAGAUGUUGCACAUCCGAACGGCAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAU ................(((((.(((((.....((....))...((((((((.....(((((....)))))...))))))))......................)))))...))))).... ( -25.30) >DroSim_CAF1 30822 120 + 1 GCUCACACUUCAUUUAGCCAGAUGUUGCACAUCCGAACGGCAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAU ................(((((.(((((.....((....))...((((((((.....(((((....)))))...))))))))......................)))))...))))).... ( -25.30) >DroEre_CAF1 24843 120 + 1 ACUCACACUUCAUUUAGCCAGAUGUUGCACAUCCGAACAGCAAGAGCAACACCAGCAGCAGCAACCUACUGAAUGUUGCUUAUUUUCACUGGCAAAAUAAAAUCAACACAAUCGGUAAAU ...........((((.(((((.(((((..(....)..))))).((((((((....(((.((....)).)))..)))))))).......))))).))))...................... ( -25.10) >DroYak_CAF1 26682 120 + 1 ACUCACACUUCAUUUAGCCAGAUGUUGCACAUCUAAACGGCAAGAGAAACACCAGAAGCAGCAACCUACUAAAUGUUGCUUAUUUUCACUGGCAAAAUAAAAUAAACACACAUGGUAAAU ...........((((.(((((.(.((((.(........))))).)((((......((((((((..........))))))))..)))).))))).))))...................... ( -22.00) >consensus ACUCACACUUCAUUUAGCCAGAUGUUGCACAUCCGAACGGCAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAU ...........((((.(((.((((.....))))..........((((((((.....(((((....)))))...)))))))).........))).))))...................... (-20.00 = -20.60 + 0.60)

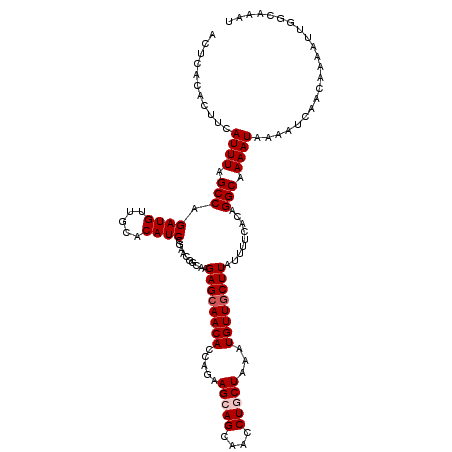
| Location | 1,400,092 – 1,400,212 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -32.04 |
| Energy contribution | -31.68 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1400092 120 - 20766785 AUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUGCCGUUCGGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGU (((..(((((...))))(((((((((.(((...(((.(..(((((((((.(((((....)))))...)))))))))..)....)))(((((.....)))))))).))))))))))..))) ( -35.40) >DroSec_CAF1 25896 120 - 1 AUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUGCCGUUCGGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGC .((..(((((...))))(((((((((.(((...(((.(..(((((((((.(((((....)))))...)))))))))..)....)))(((((.....)))))))).))))))))))..)). ( -34.80) >DroSim_CAF1 30822 120 - 1 AUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUGCCGUUCGGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGC .((..(((((...))))(((((((((.(((...(((.(..(((((((((.(((((....)))))...)))))))))..)....)))(((((.....)))))))).))))))))))..)). ( -34.80) >DroEre_CAF1 24843 120 - 1 AUUUACCGAUUGUGUUGAUUUUAUUUUGCCAGUGAAAAUAAGCAACAUUCAGUAGGUUGCUGCUGCUGGUGUUGCUCUUGCUGUUCGGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGU ((((((((((...))))(((((((((.(((((((......((((((((.((((((.......)))))))))))))).((((...........)))))).))))).))))))))))))))) ( -36.50) >DroYak_CAF1 26682 120 - 1 AUUUACCAUGUGUGUUUAUUUUAUUUUGCCAGUGAAAAUAAGCAACAUUUAGUAGGUUGCUGCUUCUGGUGUUUCUCUUGCCGUUUAGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGU ................((((((((((.(((((((......((.((((((.(((((....)))))...)))))).)).(((((((....))).)))))).))))).))))))))))..... ( -29.20) >consensus AUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUGCCGUUCGGAUGUGCAACAUCUGGCUAAAUGAAGUGUGAGU .................(((((((((.(((...(((.(..(((((((((.(((((....)))))...)))))))))..)....)))(((((.....)))))))).)))))))))...... (-32.04 = -31.68 + -0.36)

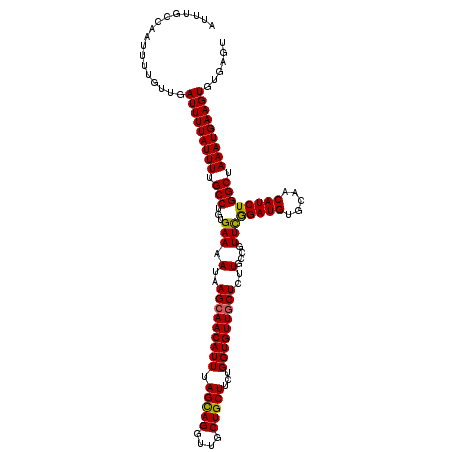
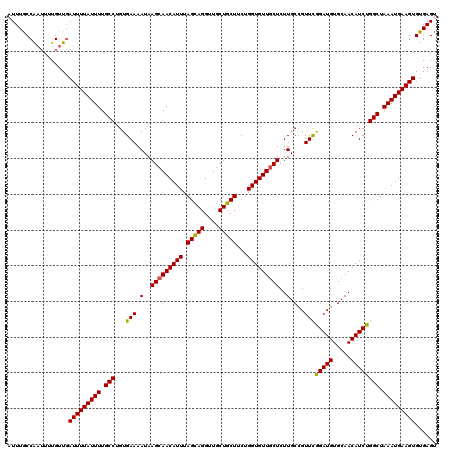
| Location | 1,400,132 – 1,400,252 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1400132 120 + 20766785 CAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUAG ....((((((.....((((.(((..((((....(((((.((((((((....).)))))))...))))).....((.(((((.........))))).)))))).)))))))..)))))).. ( -26.90) >DroSec_CAF1 25936 120 + 1 CAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUAG ....((((((.....((((.(((..((((....(((((.((((((((....).)))))))...))))).....((.(((((.........))))).)))))).)))))))..)))))).. ( -26.90) >DroSim_CAF1 30862 120 + 1 CAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUAG ....((((((.....((((.(((..((((....(((((.((((((((....).)))))))...))))).....((.(((((.........))))).)))))).)))))))..)))))).. ( -26.90) >DroEre_CAF1 24883 120 + 1 CAAGAGCAACACCAGCAGCAGCAACCUACUGAAUGUUGCUUAUUUUCACUGGCAAAAUAAAAUCAACACAAUCGGUAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUUG ...(((((((...(((((((.((......))..)))))))..........(((....................((((((((.........))))))))((....)))))...))))))). ( -28.40) >DroYak_CAF1 26722 120 + 1 CAAGAGAAACACCAGAAGCAGCAACCUACUAAAUGUUGCUUAUUUUCACUGGCAAAAUAAAAUAAACACACAUGGUAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUAG .....((((......((((((((..........))))))))..)))).(((((((..(((.............((((((((.........))))))))((....))..)))..))))))) ( -25.80) >consensus CAAGAGCAACACCAGAAGCAGCAACCUGCUAAAUGUUGCUUAUUUUCACAGGCAAAAUAAAAUCAACAAAAUUGGCAAAUUGUUUAAUUGAAUUUACCGCAGAUGCGCUUAAGUUGCUAG ....((((((.....(((((((((..((((((.(((((.((((((((....).)))))))...)))))...))))))..)))))..............((....))))))..)))))).. (-23.80 = -23.80 + 0.00)

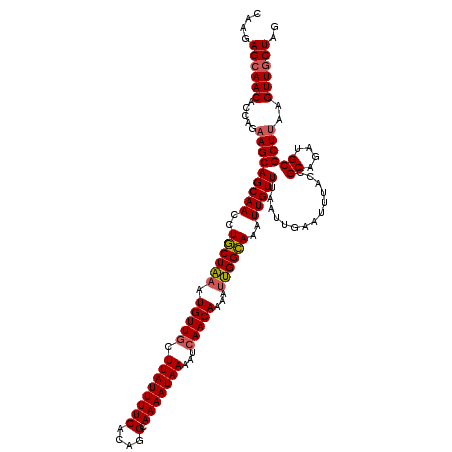
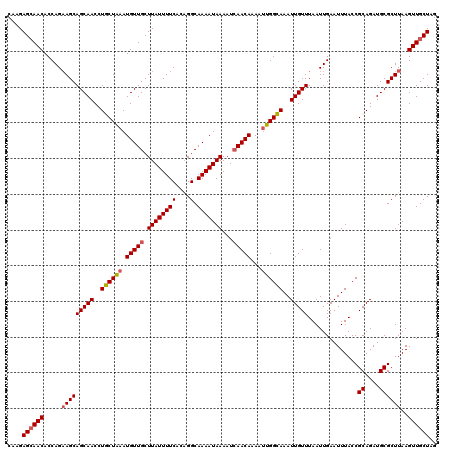
| Location | 1,400,132 – 1,400,252 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.78 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1400132 120 - 20766785 CUAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUG ..((((((..((((((((((((((((((((.........)))))))).....(((((...(((((((.......))))))).)))))....))))).))).)))).....)))))).... ( -33.70) >DroSec_CAF1 25936 120 - 1 CUAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUG ..((((((..((((((((((((((((((((.........)))))))).....(((((...(((((((.......))))))).)))))....))))).))).)))).....)))))).... ( -33.70) >DroSim_CAF1 30862 120 - 1 CUAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUG ..((((((..((((((((((((((((((((.........)))))))).....(((((...(((((((.......))))))).)))))....))))).))).)))).....)))))).... ( -33.70) >DroEre_CAF1 24883 120 - 1 CAAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUACCGAUUGUGUUGAUUUUAUUUUGCCAGUGAAAAUAAGCAACAUUCAGUAGGUUGCUGCUGCUGGUGUUGCUCUUG ..((((((...((((((....(((((((((.........)))))))))..))))))...........((((((.......((((((.........))))))...)))))))))))).... ( -35.40) >DroYak_CAF1 26722 120 - 1 CUAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUACCAUGUGUGUUUAUUUUAUUUUGCCAGUGAAAAUAAGCAACAUUUAGUAGGUUGCUGCUUCUGGUGUUUCUCUUG ..((((.(..((((((((((((((((((((.........))))))))((((.(((((((((((((.....)))).)))))))))))))...))))).))).))))..).))))....... ( -29.50) >consensus CUAGCAACUUAAGCGCAUCUGCGGUAAAUUCAAUUAAACAAUUUGCCAAUUUUGUUGAUUUUAUUUUGCCUGUGAAAAUAAGCAACAUUUAGCAGGUUGCUGCUUCUGGUGUUGCUCUUG ..((((((..((((((((((((((((((((.........)))))))).....(((((...(((((((.......))))))).)))))....))))).))).)))).....)))))).... (-30.66 = -30.78 + 0.12)

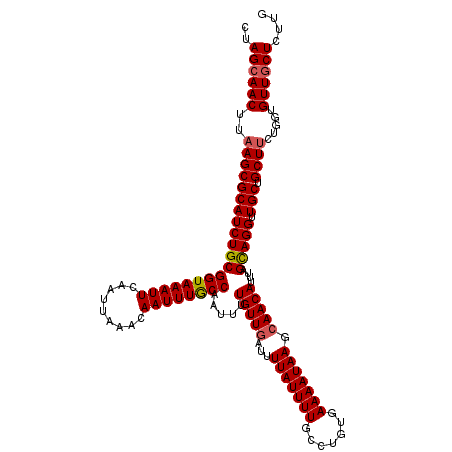
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:10 2006