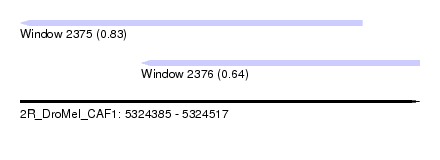
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,324,385 – 5,324,517 |
| Length | 132 |
| Max. P | 0.827441 |

| Location | 5,324,385 – 5,324,498 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.05 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5324385 113 - 20766785 CACUCCUCAUUC--AAUCC-CC---AGCUUCCUUGAAGAACUUCGGUGACAAGACCGCCAACCUCUUCAGCAAGAAGAAGGAUGAGGCCGAGAAGCUGGCCAACGAGAAGGCUGCCGAG ............--..((.-.(---(((((.((((.......((((((.......))))..(((((((.....)))).)))..))(((((......)))))..)))).))))))..)). ( -32.80) >DroVir_CAF1 643 115 - 1 UACUUGUUUUGCUAUUUCG-GC---AGCCUCCCUGAAGAGCUUUGGCGACAAGACCGUCAACUUGUUCAGCAAGAAGAAGGAUGAGGCCGAGAAGUUGGCCAGCGACAAGGCUGUCGAG ................(((-((---(((((.(((........((((((.......))))))((((.....))))....)))....((((((....)))))).......)))))))))). ( -38.00) >DroGri_CAF1 972 103 - 1 CUAU------------UCG-AC---AGCGUCCCUGAAGAGCUUUGGCGACAAGACCGCCAAUCUGUUCAGCAGGAAGAAGGACGAGGCCGAGAAGUUGGCCAGCGACAAAUCUGUCGAG ....------------(((-((---((((((((((((.((..((((((.......)))))).)).))))).........))))).((((((....))))))..........))))))). ( -41.60) >DroWil_CAF1 644 115 - 1 CUUUCAUUUUUCU-AAUCCUUU---AGCCUCUUUGAAGAACUUUGGCGACAAGACAGCCAAUCUGUUCAGCAAGAAGAAGGAUGAGGCUGAGAAAUUGGCCAACGAUAAGGCCACCGAG ...((...(((((-...((((.---..(((((((...((((.(((((.........)))))...)))).....)))).)))..))))...))))).(((((........)))))..)). ( -33.40) >DroMoj_CAF1 672 115 - 1 CUAUUAUAUUUCUUUUCCG-AC---AGCCUCUCUGAAGAGCUUUGGCGACAAGACCGCCAACUUGUUCAGCAAGAAGAAGGAUGAGGCCGAGAAGCUGGCCAACGAAAAGGCAGCCGAG ..........(((((((..-.(---(.((((((((..((((.((((((.......))))))...))))..).)).)).))).)).(((((......)))))...)))))))........ ( -31.60) >DroAna_CAF1 642 115 - 1 -CCUUUUCGUUU--AUUUC-UCGGUAGCAUCCUUGAAGAACUUUGGCGACAAGACGGCCAAUCUGUUCAGCAAGAAGAAGGAUGAGGCUGAGAAGCUGGCCAACGAGAAGGCUAACGAG -(((((((((((--(((((-(((((..((((((((..((((.(((((.........)))))...))))..))......))))))..))))))))).))...))))))))))........ ( -41.30) >consensus CUCUUAUCUUUC__AUUCC_AC___AGCCUCCCUGAAGAACUUUGGCGACAAGACCGCCAACCUGUUCAGCAAGAAGAAGGAUGAGGCCGAGAAGCUGGCCAACGACAAGGCUGCCGAG ..........................((((.((((..((((.((((((.......))))))...))))..))))...........(((((......))))).......))))....... (-22.44 = -22.72 + 0.28)


| Location | 5,324,425 – 5,324,517 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -19.49 |
| Consensus MFE | -13.89 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5324425 92 - 20766785 ACAGUC-AGGAUCUAAAAU------GCACUCCUCAUUC--AAUCCCCAGCUUCCUUGAAGAACUUCGGUGACAAGACCGCCAACCUCUUCAGCAAGAAGAA ......-.((((....(((------(.......)))).--.))))....((((.(((((((..((.((((.......))))))..)))))))...)))).. ( -15.60) >DroSec_CAF1 1097 92 - 1 CCAGUC-AGGAUCUAAAAU------GCUCUCCUCCUUC--AAUCCCCAGCUUCCUUGAAGAACUUCGGCGACAAGACCGCCAACCUCUUCAGCAAGAAGAA ......-((((........------....)))).....--.........((((.(((((((..((.((((.......))))))..)))))))...)))).. ( -16.70) >DroSim_CAF1 1133 92 - 1 CCAGUC-AGGAUCUAAAAU------GAACUCCUCAUUC--AAUCCCCAGCUUCCUUGAAGAACUUCGGCGACAAGACCGCCAACCUCUUCAGCAAGAAGAA ......-.((((....(((------((.....))))).--.))))....((((.(((((((..((.((((.......))))))..)))))))...)))).. ( -20.20) >DroEre_CAF1 1073 95 - 1 CUAGUC-AGGAUCCACAAUAAC---GCACUCCUUGUUC--AAUCCCCAGCCUCGUUGAAGAACUUCGGCGACAAGACCGCCAACCUCUUCAGCAAGAAGAA ....((-.((((..((((....---.......))))..--.)))).....((.((((((((..((.((((.......))))))..)))))))).))..)). ( -20.20) >DroYak_CAF1 1183 91 - 1 UCAAGC-AGGAUCUACAAU------GCACUCCUC-UUU--AAUCCGCAGCUUCGUUGAAGAACUUCGGCGACAAGACCGCCAACCUCUUCAGCAAGAAGAA ....((-.((((.......------.........-...--.))))))..((((((((((((..((.((((.......))))))..))))))))..)))).. ( -22.15) >DroMoj_CAF1 712 98 - 1 UGAA--UUGGAAGUAAAAUUGAUUU-CUAUUAUAUUUCUUUUCCGACAGCCUCUCUGAAGAGCUUUGGCGACAAGACCGCCAACUUGUUCAGCAAGAAGAA ....--(((((((..((((((((..-..)))).))))..)))))))....((..(((((.((..((((((.......)))))).)).)))))..))..... ( -22.10) >consensus CCAGUC_AGGAUCUAAAAU______GCACUCCUCAUUC__AAUCCCCAGCUUCCUUGAAGAACUUCGGCGACAAGACCGCCAACCUCUUCAGCAAGAAGAA .................................................((((.(((((((..((.((((.......))))))..)))))))...)))).. (-13.89 = -14.12 + 0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:31 2006