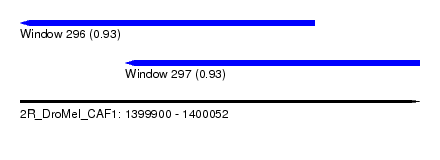
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,399,900 – 1,400,052 |
| Length | 152 |
| Max. P | 0.933438 |

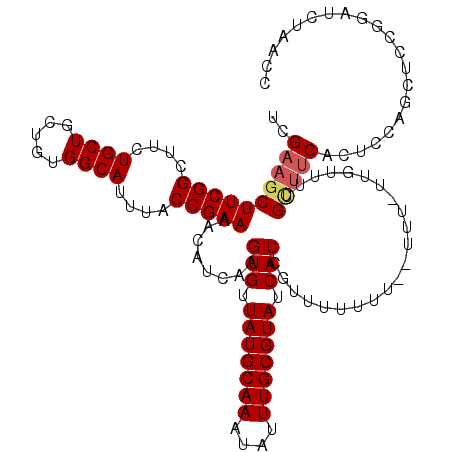
| Location | 1,399,900 – 1,400,012 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1399900 112 - 20766785 CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU--------GUUUUGUCCCACUCCAGCUCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUAAGUCGACAGUGAAAUUU .((((((((.(((((((....))))))).............--------(((..((((............))))..)))))))))))...(((((((.(.......).)))))))..... ( -23.70) >DroSec_CAF1 25696 120 - 1 CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUUUUUUUAUUUUUUUGCUUCACUCCAGCUCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUAAGUCGACAGUGAAAUUU ......(((.(((((((....))))))).)))........................((((((.((((...(((((.........)))))...))))..(((......))))))))).... ( -22.20) >DroSim_CAF1 30624 118 - 1 CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUU--UUUUUAUUUUUUUGCUUCACUCCAGCUCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUAAGUCGACAGUGAAAUUU ......(((.(((((((....))))))).))).......--...............((((((.((((...(((((.........)))))...))))..(((......))))))))).... ( -22.20) >DroEre_CAF1 24645 118 - 1 CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU--UUUUUUGUUGUGGUUCACUCCAGCUCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUUAGUCCACAGUGAAAUUU ......(((.(((((((....))))))).))).........--..(((..((((((...(((.((((...(((((.........)))))...)))).)))......))))))..)))... ( -27.60) >DroYak_CAF1 26500 102 - 1 CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGCUUUUUU--U----------GCUUCAC------UCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUUAGUCGACAGUGAAAUUU .((((((((....(((((.....(((......)))....))--)----------))..)))------...((......))..)))))...(((((((.(.......).)))))))..... ( -22.20) >consensus CAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU__UUU_UUGUUUUGCUUCACUCCAGCUCCGGAUCUAACCAUUUGAUUUAUCGCUGUAGUCUAAGUCGACAGUGAAAUUU .((((((((.(((((((....))))))).................................(((......)))......))))))))...(((((((.(.......).)))))))..... (-19.92 = -20.52 + 0.60)

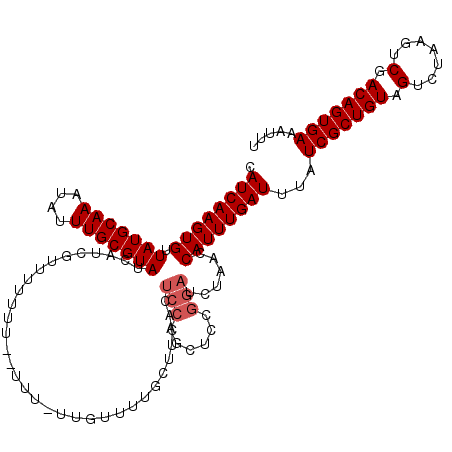
| Location | 1,399,940 – 1,400,052 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1399940 112 - 20766785 UCGAAGCUUCGGCUUCUGCUGCUGUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU--------GUUUUGUCCCACUCCAGCUCCGGAUCUAACC ..(((((....))))).((((..((((.((..(.((((((.((...(((.(((((((....))))))).))).)).)))))--------).)..)).))))..))))............. ( -32.40) >DroSec_CAF1 25736 120 - 1 UCGAAGCUUCGGCUUCUGCUGCUGUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUUUUUUUAUUUUUUUGCUUCACUCCAGCUCCGGAUCUAACC ..(((((....))))).((((..((((((......((.....))..(((.(((((((....))))))).))).....................)))..)))..))))............. ( -27.40) >DroSim_CAF1 30664 118 - 1 UCGAAGCUUCGGCUUCUGCUGCUGUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUU--UUUUUAUUUUUUUGCUUCACUCCAGCUCCGGAUCUAACC ..(((((....))))).((((..((((((......(((((......(((.(((((((....))))))).))).......--))))).......)))..)))..))))............. ( -28.04) >DroEre_CAF1 24685 118 - 1 UCGAAGCUUCGGCUUCUGCUGCUCUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU--UUUUUUGUUGUGGUUCACUCCAGCUCCGGAUCUAACC ..(((((....)))))((((.....))))....((((((((.....(((.(((((((....))))))).))).)))))...--......((((..(....)..))))..)))........ ( -28.70) >DroYak_CAF1 26540 102 - 1 UCGAAGCUUCGGCUUGUGCUGCUGUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGCUUUUUU--U----------GCUUCAC------UCCGGAUCUAACC ..((((((((((...(((((.....)))))...)))))......(((((.(((((((....)))))))....)))))....--.----------)))))..------............. ( -28.30) >consensus UCGAAGCUUCGGCUUCUGCUGCUGUGGCAUUUACCGAAAACAUCAAGUGUUAUGCAAAUAUUUGCGUAUCAUCGUUUUUUU__UUU_UUGUUUUGCUUCACUCCAGCUCCGGAUCUAACC ..((((((((((....((((.....))))....)))))........(((.(((((((....))))))).)))......................)))))..................... (-21.26 = -21.74 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:05 2006