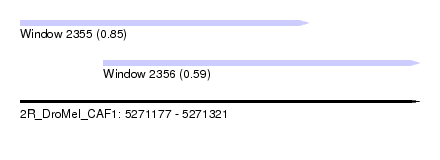
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,271,177 – 5,271,321 |
| Length | 144 |
| Max. P | 0.845395 |

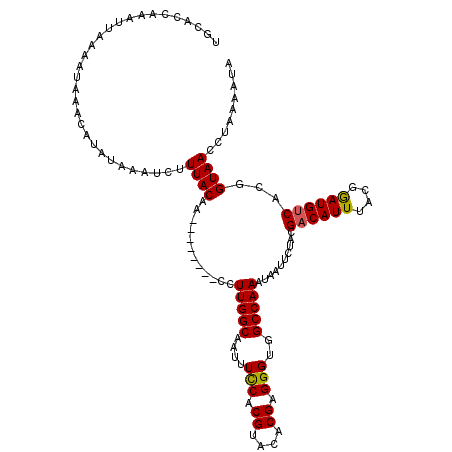
| Location | 5,271,177 – 5,271,281 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -11.37 |
| Consensus MFE | -10.59 |
| Energy contribution | -10.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5271177 104 + 20766785 -ACCAAAAAUAUAAACGCUUACUAAGACAUC-----------AAAUUAAAAUAAACCUAUAAAUCUUUACAAUCUUAAGUCCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAU -..............................-----------........................................(((((....(((.((....)).)))..))))).. ( -11.30) >DroSec_CAF1 139020 107 + 1 AAACAAAAACAUAAACAU-AAACAAGACAUCAUUUUUGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA--------CCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAU ..................-...(((((......)))))..................................--------..(((((....(((.((....)).)))..))))).. ( -11.90) >DroSim_CAF1 127253 107 + 1 AAACAAAAACAUAAACAU-AAACAAGACAUCAUUUUUGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA--------CCUUGGCAAUUUUCACGUACACGAGGGUGGCCAAAU ..................-...(((((......)))))..................................--------..(((((.((((((........)))))).))))).. ( -10.90) >consensus AAACAAAAACAUAAACAU_AAACAAGACAUCAUUUUUGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA________CCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAU ..................................................................................(((((....(((.((....)).)))..))))).. (-10.59 = -10.37 + -0.22)

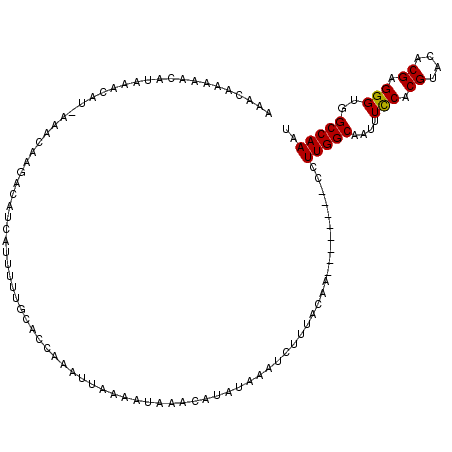
| Location | 5,271,207 – 5,271,321 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5271207 114 + 20766785 ------AAAUUAAAAUAAACCUAUAAAUCUUUACAAUCUUAAGUCCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAUAAUUUCACGACAUUUAAGAAUGACGCGGUAACGUAAAACA ------...........................((.(((((((((.(((((....(((.((....)).)))..)))))..........)))..)))))).))..(((....)))...... ( -17.70) >DroSec_CAF1 139055 112 + 1 UGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA--------CCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAUAAUUCUACGACAUUUACGGAUGUCACGGUAACCUAAAAUA ...(((..............................--------..(((((....(((.((....)).)))..)))))..........((((((....))))))..)))........... ( -20.80) >DroSim_CAF1 127288 112 + 1 UGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA--------CCUUGGCAAUUUUCACGUACACGAGGGUGGCCAAAUAAUUCUACGACAUUCACGGAUGUCACGGUAACCUAAAAUA ...(((..............................--------..(((((.((((((........)))))).)))))..........((((((....))))))..)))........... ( -20.90) >consensus UGCACCAAAUUAAAAUAAACAUAUAAAUCUUUACAA________CCUUGGCAAUUUCCACGUACACGAGGGUGGCCAAAUAAUUCUACGACAUUUACGGAUGUCACGGUAACCUAAAAUA ..............................((((............(((((....(((.((....)).)))..)))))..........((((((....))))))...))))......... (-16.94 = -16.83 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:12 2006