
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,255,658 – 5,255,749 |
| Length | 91 |
| Max. P | 0.825798 |

| Location | 5,255,658 – 5,255,749 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -20.05 |
| Consensus MFE | -10.28 |
| Energy contribution | -10.20 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5255658 91 + 20766785 CUCG----------UGUCGCUCCGGAUUCUUUUCAUUUUUGGAAUCGCCCGACGCUCGGCAUUUUUCAUGCAACGAGGAUUUUUCCCAUCUCAAUCCCCCA ((((----------(((((.....((((((..........))))))...))))))...((((.....))))...)))........................ ( -18.30) >DroSec_CAF1 123669 89 + 1 UUCG----------UGUCGCUCCGGAUUCUUUUCAUUUUUGGAAUCGCCCGACGCUCGGCAUUUUUCAUGCAACGAGGA-UUUUCCCAUCUCAAUCCC-CA ...(----------(((((.....((((((..........))))))...))))))(((((((.....))))..)))(((-((..........))))).-.. ( -18.10) >DroSim_CAF1 111059 90 + 1 UUCG----------UGUCGCUCCGGAUUCUUUUCAUUUUUGGAAUCGCCCGACACUCGGCAUUUUUCAUGCAACGAGGACUUUUCCCAUCUCAAUCCC-CA ...(----------(((((.....((((((..........))))))...))))))(((((((.....))))..)))(((....)))............-.. ( -17.50) >DroEre_CAF1 124316 76 + 1 CUGA----------UGUCGCACCGGGUUCUUUUCAUUUUUGAAAUCGCCCGACGCUCGGCAUUUUUCAUGCUACGAGGAUUUUUCC--------------- ..((----------.(((....(((((...(((((....)))))..)))))...((((((((.....))))..)))))))...)).--------------- ( -19.80) >DroYak_CAF1 125554 76 + 1 CUCG----------UGUCGCACCGGGUUCUUUUCAUUUUUGGAAUCGCCCGACGCUCGGCAUUUUUCAUGCUACGACGAUUUUCCC--------------- ....----------((((((..(((((...(((((....)))))..)))))..))..(((((.....)))))..))))........--------------- ( -21.20) >DroAna_CAF1 139753 97 + 1 UGCGAAAGUCUGUCUUCCGCUUCGGGCGCCUUUCAUUUUUGGAAUCGCCCGACACUCAACAUUUUUCAUGUUAUAUGCAUUUGGCCAAACGCCUUG----G .(((.(((.....))).))).(((((((..(((((....))))).))))))).....(((((.....))))).....((...(((.....))).))----. ( -25.40) >consensus CUCG__________UGUCGCUCCGGAUUCUUUUCAUUUUUGGAAUCGCCCGACGCUCGGCAUUUUUCAUGCAACGAGGAUUUUUCCCAUCUCAAUC____A ..............(((((....((.....(((((....)))))...)))))))((((((((.....))))..))))........................ (-10.28 = -10.20 + -0.08)

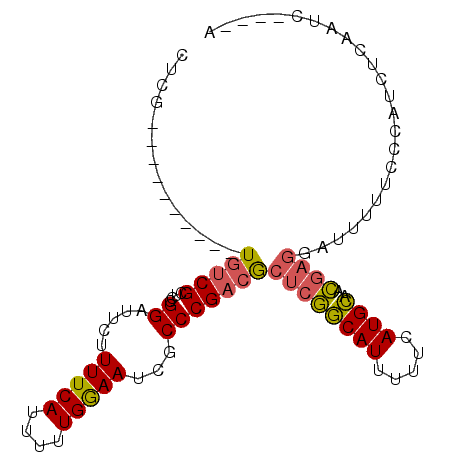
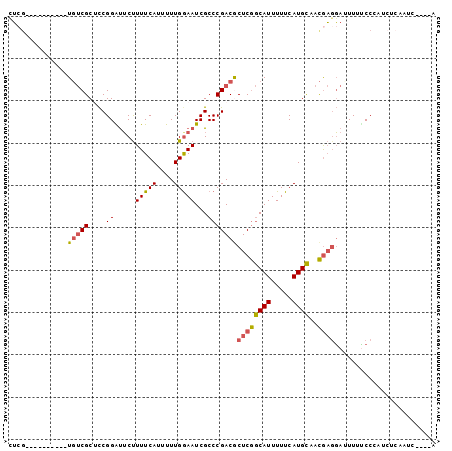
| Location | 5,255,658 – 5,255,749 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -12.48 |
| Energy contribution | -11.35 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5255658 91 - 20766785 UGGGGGAUUGAGAUGGGAAAAAUCCUCGUUGCAUGAAAAAUGCCGAGCGUCGGGCGAUUCCAAAAAUGAAAAGAAUCCGGAGCGACA----------CGAG ..(((((((...........)))))))(((((.........(((........)))(((((............)))))....))))).----------.... ( -22.00) >DroSec_CAF1 123669 89 - 1 UG-GGGAUUGAGAUGGGAAAA-UCCUCGUUGCAUGAAAAAUGCCGAGCGUCGGGCGAUUCCAAAAAUGAAAAGAAUCCGGAGCGACA----------CGAA .(-((((((..........))-)))))(((((.........(((........)))(((((............)))))....))))).----------.... ( -22.10) >DroSim_CAF1 111059 90 - 1 UG-GGGAUUGAGAUGGGAAAAGUCCUCGUUGCAUGAAAAAUGCCGAGUGUCGGGCGAUUCCAAAAAUGAAAAGAAUCCGGAGCGACA----------CGAA .(-((((((...........)))))))...((((.....))))...((((((..((((((............)))))..)..)))))----------)... ( -22.70) >DroEre_CAF1 124316 76 - 1 ---------------GGAAAAAUCCUCGUAGCAUGAAAAAUGCCGAGCGUCGGGCGAUUUCAAAAAUGAAAAGAACCCGGUGCGACA----------UCAG ---------------(((....)))(((..((((.....)))))))((..((((...(((((....)))))....))))..))....----------.... ( -20.70) >DroYak_CAF1 125554 76 - 1 ---------------GGGAAAAUCGUCGUAGCAUGAAAAAUGCCGAGCGUCGGGCGAUUCCAAAAAUGAAAAGAACCCGGUGCGACA----------CGAG ---------------.......((((.((.((((.....))))...((..(((((..(((.......)))..)..))))..)).)))----------))). ( -18.60) >DroAna_CAF1 139753 97 - 1 C----CAAGGCGUUUGGCCAAAUGCAUAUAACAUGAAAAAUGUUGAGUGUCGGGCGAUUCCAAAAAUGAAAGGCGCCCGAAGCGGAAGACAGACUUUCGCA .----...(((.....))).....(((.((((((.....)))))).)))(((((((.(((.......)))...))))))).(((((((.....))))))). ( -32.90) >consensus U____GAUUGAGAUGGGAAAAAUCCUCGUAGCAUGAAAAAUGCCGAGCGUCGGGCGAUUCCAAAAAUGAAAAGAACCCGGAGCGACA__________CGAA .........................(((..((((.....)))))))((..(((((..(((.......)))..)..))))..)).................. (-12.48 = -11.35 + -1.13)

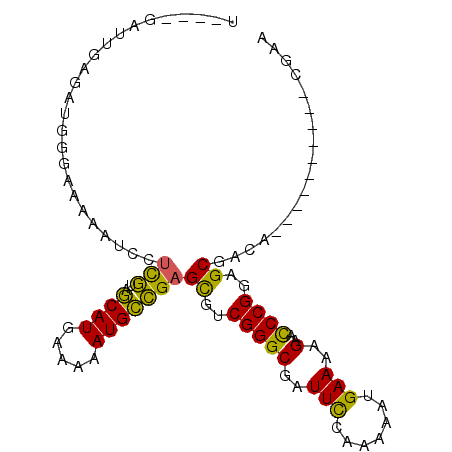
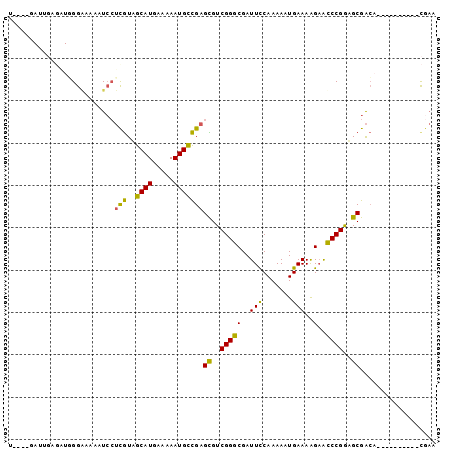
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:08 2006