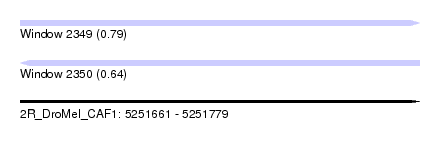
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,251,661 – 5,251,779 |
| Length | 118 |
| Max. P | 0.791583 |

| Location | 5,251,661 – 5,251,779 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.53 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -6.50 |
| Energy contribution | -6.50 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5251661 118 + 20766785 UACAGAAGAUUGAA-UUACAAGGAUGCAAUUGCCUGGACAAGAGAUAUCUCCCAUCUUUAUGGCAUACCAUCCAAGCGUUUAAUCUUUAAGUGAUCAGG-ACUUCUUCCUGUGCACUACG ....((((((((((-(.....(((((....((((.(((...(((....)))...)))....))))...)))))....))))))))))).((((..((((-(.....)))))..))))... ( -35.90) >DroSec_CAF1 119772 113 + 1 UACAGAAAUAUAGA-U-----CCAGGCAAUUGCCUGGACAAGAGAUAUCUCCCAUCUUUACGGCAUACCAUCCAACCGUUUAAUCUUUAAGUGAUCAGG-ACUUUUUCCUGUGCACUUCG ...(((.....(((-(-----((((((....)))))))...(((....)))...)))..((((............))))....)))..(((((..((((-(.....)))))..))))).. ( -28.40) >DroSim_CAF1 107165 113 + 1 UACAGAAAUAUAGA-U-----CCAGGCAAUUGCCUGGACAAGAGAUAUCUCCCAUCUUUACGGCAUACCAUCCAACCGUUUAAUCUUUAAGUGAUCAGG-ACUUCUUCCUGUGCACGUCG ....((..((.(((-(-----((((((....)))))))..((((((.......))))))((((............))))....))).)).(((..((((-(.....)))))..))).)). ( -27.90) >DroEre_CAF1 120396 99 + 1 UUUACA---AUAGAAGUACGUG-------------GUACAAGUGAUAUCUCCCAUCUUUAUGGCAUCG----CAACCGUUUAAUCCUCAAGUGAUCAGG-ACUUCUUCCUGUGCACUCUG ......---...((..((..((-------------((....(((((.....((((....)))).))))----).))))..))....)).((((..((((-(.....)))))..))))... ( -17.70) >DroYak_CAF1 121558 97 + 1 UACACAC-UAUAGAAGUACGAGAAUGCAAUUGCUUGCACAAGUGGUAUCACCCAUCUU----------------------UGAUCUUCAAGGGAUCAGGCAUUUUUUUCUGUGCACUCUG .......-(((((((....((((((((.((..(((....)))..))...........(----------------------((((((.....))))))))))))))))))))))....... ( -22.70) >consensus UACAGAA_UAUAGA_UUAC__GCAGGCAAUUGCCUGGACAAGAGAUAUCUCCCAUCUUUACGGCAUACCAUCCAACCGUUUAAUCUUUAAGUGAUCAGG_ACUUCUUCCUGUGCACUCCG .........................................(((....))).......................................(((..((((........))))..))).... ( -6.50 = -6.50 + -0.00)

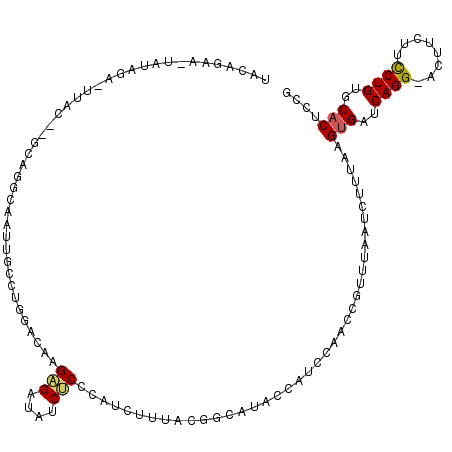
| Location | 5,251,661 – 5,251,779 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.53 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -11.44 |
| Energy contribution | -12.44 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5251661 118 - 20766785 CGUAGUGCACAGGAAGAAGU-CCUGAUCACUUAAAGAUUAAACGCUUGGAUGGUAUGCCAUAAAGAUGGGAGAUAUCUCUUGUCCAGGCAAUUGCAUCCUUGUAA-UUCAAUCUUCUGUA ...((((..(((((.....)-))))..))))..((((((....(((((.((((....))))...((..((((....))))..)))))))(((((((....)))))-)).))))))..... ( -35.90) >DroSec_CAF1 119772 113 - 1 CGAAGUGCACAGGAAAAAGU-CCUGAUCACUUAAAGAUUAAACGGUUGGAUGGUAUGCCGUAAAGAUGGGAGAUAUCUCUUGUCCAGGCAAUUGCCUGG-----A-UCUAUAUUUCUGUA ..(((((..(((((.....)-))))..)))))..(((((..(((((..........)))))...((..((((....))))..))(((((....))))))-----)-)))........... ( -36.50) >DroSim_CAF1 107165 113 - 1 CGACGUGCACAGGAAGAAGU-CCUGAUCACUUAAAGAUUAAACGGUUGGAUGGUAUGCCGUAAAGAUGGGAGAUAUCUCUUGUCCAGGCAAUUGCCUGG-----A-UCUAUAUUUCUGUA ....(((..(((((.....)-))))..)))....(((((..(((((..........)))))...((..((((....))))..))(((((....))))))-----)-)))........... ( -34.40) >DroEre_CAF1 120396 99 - 1 CAGAGUGCACAGGAAGAAGU-CCUGAUCACUUGAGGAUUAAACGGUUG----CGAUGCCAUAAAGAUGGGAGAUAUCACUUGUAC-------------CACGUACUUCUAU---UGUAAA .....((((..(((((.(((-(((.(.....).))))))....(((.(----(((..((((....))))((....))..))))))-------------).....)))))..---)))).. ( -21.80) >DroYak_CAF1 121558 97 - 1 CAGAGUGCACAGAAAAAAAUGCCUGAUCCCUUGAAGAUCA----------------------AAGAUGGGUGAUACCACUUGUGCAAGCAAUUGCAUUCUCGUACUUCUAUA-GUGUGUA ..((((((..((((.........(((((.......)))))----------------------...(..((((....))))..)((((....)))).)))).)))))).....-....... ( -21.00) >consensus CGAAGUGCACAGGAAGAAGU_CCUGAUCACUUAAAGAUUAAACGGUUGGAUGGUAUGCCAUAAAGAUGGGAGAUAUCUCUUGUCCAGGCAAUUGCAUGC__GUAA_UCUAUA_UUCUGUA ..(((((..((((........))))..)))))................................((..((((....))))..)).................................... (-11.44 = -12.44 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:06 2006