
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,233,425 – 5,233,526 |
| Length | 101 |
| Max. P | 0.969597 |

| Location | 5,233,425 – 5,233,526 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -21.36 |
| Energy contribution | -22.55 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
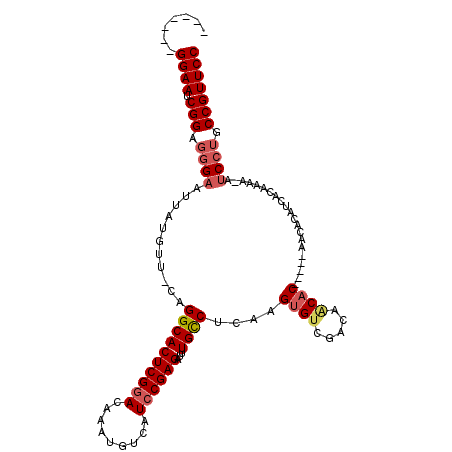
>2R_DroMel_CAF1 5233425 101 + 20766785 CAGAAUUGGAAUCGGAGGGAAUUAUGUU-CAGGCACUCGGACAAAUGUCAUCCGAGAUUUGCCUCAAGUGUCGACAACACAUC----------ACAAAA-AUCCUGCCGUUCC .......((((.(((.((((.((.(((.-.(((((((((((.........))))))...)))))...((((.....))))...----------))).))-.)))).))))))) ( -30.90) >DroSec_CAF1 108968 111 + 1 CAGAAUUGGAAUCGGAGGGAAUUAUGUU-CAGGCACUCGGACAAAUGUCAUCCGAGAUUUGUCUCAAGUGUCGACAACACAUCGCAACACAACACAAAA-AUCCUGCCGUUCC .......((((.(((.((((.((.(((.-..((((((.((((((((.((....)).))))))).).))))))((.......))..........))).))-.)))).))))))) ( -32.10) >DroEre_CAF1 109840 100 + 1 -------GGAAUCGGAGGGAAUUAUGUUCCAGGCACUCGGACAAAUGUCAUCCGAGAUUUGCCACAAGUGUCGAUAGCCC-----AAUACAUCACAAAA-AUCCUGCCGUUCC -------((((.(((.((((.((.(((....((((((((((.........))))))...))))....((((.(......)-----.))))...))).))-.)))).))))))) ( -29.30) >DroYak_CAF1 110615 100 + 1 -------GGAAUCGGAGAGAAUUAUGUU-CAGGCACUCGAACAAAUGUCAUCCGAGAUUUGCCUCAAGUGUCGAUACCAC-----AACACAUCACAAAAAACCCGGCCGUUCC -------((((.(((...((((...)))-).((((((.((.(((((.((....)).)))))..)).))))))........-----.....................))))))) ( -22.60) >consensus _______GGAAUCGGAGGGAAUUAUGUU_CAGGCACUCGGACAAAUGUCAUCCGAGAUUUGCCUCAAGUGUCGACAACAC_____AACACAUCACAAAA_AUCCUGCCGUUCC .......((((.(((.((((...........((((((((((.........))))))...))))....((((.....)))).....................)))).))))))) (-21.36 = -22.55 + 1.19)


| Location | 5,233,425 – 5,233,526 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
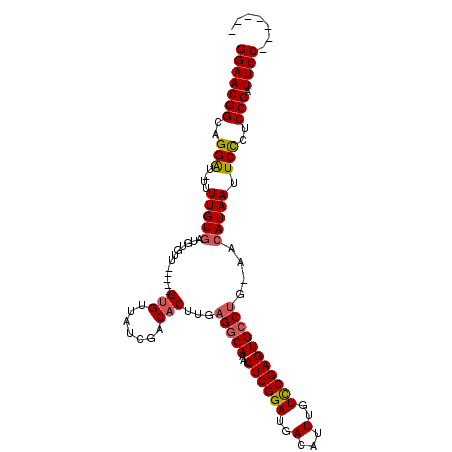
>2R_DroMel_CAF1 5233425 101 - 20766785 GGAACGGCAGGAU-UUUUGU----------GAUGUGUUGUCGACACUUGAGGCAAAUCUCGGAUGACAUUUGUCCGAGUGCCUG-AACAUAAUUCCCUCCGAUUCCAAUUCUG (((((((..(((.-..((((----------(..(((((...)))))((.(((((...((((((..(...)..))))))))))).-))))))).)))..))).))))....... ( -37.00) >DroSec_CAF1 108968 111 - 1 GGAACGGCAGGAU-UUUUGUGUUGUGUUGCGAUGUGUUGUCGACACUUGAGACAAAUCUCGGAUGACAUUUGUCCGAGUGCCUG-AACAUAAUUCCCUCCGAUUCCAAUUCUG (((((((..(((.-..(((((((......((((.....)))).((((((.(((((((.((....)).)))))))))))))....-))))))).)))..))).))))....... ( -36.70) >DroEre_CAF1 109840 100 - 1 GGAACGGCAGGAU-UUUUGUGAUGUAUU-----GGGCUAUCGACACUUGUGGCAAAUCUCGGAUGACAUUUGUCCGAGUGCCUGGAACAUAAUUCCCUCCGAUUCC------- (((((((..(((.-..(((((..((.((-----((....)))).))....((((...((((((..(...)..)))))))))).....))))).)))..))).))))------- ( -31.40) >DroYak_CAF1 110615 100 - 1 GGAACGGCCGGGUUUUUUGUGAUGUGUU-----GUGGUAUCGACACUUGAGGCAAAUCUCGGAUGACAUUUGUUCGAGUGCCUG-AACAUAAUUCUCUCCGAUUCC------- (((((((..(((....(((((..(((((-----(......))))))((.(((((...((((((..(...)..))))))))))).-)))))))..))).))).))))------- ( -33.30) >consensus GGAACGGCAGGAU_UUUUGUGAUGUGUU_____GUGUUAUCGACACUUGAGGCAAAUCUCGGAUGACAUUUGUCCGAGUGCCUG_AACAUAAUUCCCUCCGAUUCC_______ (((((((..(((....(((((............(((.......)))...(((((...((((((..(...)..)))))))))))....))))).)))..))).))))....... (-26.76 = -27.20 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:00 2006