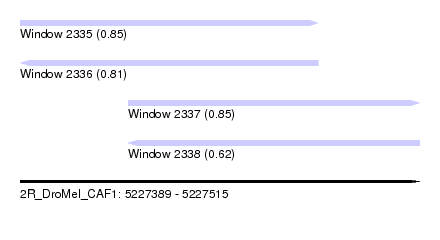
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,227,389 – 5,227,515 |
| Length | 126 |
| Max. P | 0.849436 |

| Location | 5,227,389 – 5,227,483 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
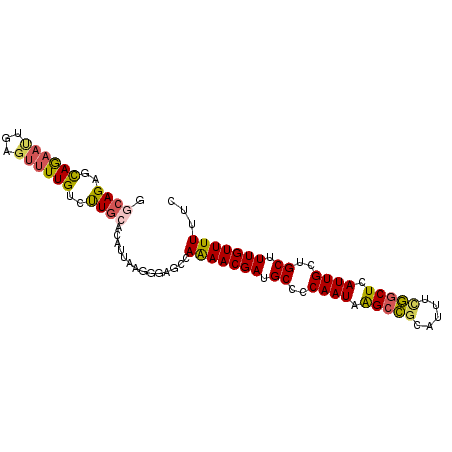
>2R_DroMel_CAF1 5227389 94 + 20766785 GGGAGAGUAGAAUUGAGUUUUGUCUUGCAUAUUAAGGGCGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUGC (.((((.((((((...)))))))))).)...........((.(((((((.((..((((.(((((.....))))).))))..)).))))))).)) ( -24.90) >DroSec_CAF1 103132 94 + 1 GCCAGAGCAGAAUUGAGUUUUGUCUUGCACAUUAAGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUGC ..((((((((......((((((.(((.(.......).))).)))))).......((((.(((((.....))))).))))))))))))....... ( -29.00) >DroSim_CAF1 89293 94 + 1 GGCAGAGCAGAAUUGAGUUUUGUCUUGCACAUUACGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUAC ((((((((((......((((((.(((.(.......).))).)))))).......((((.(((((.....))))).))))))))))))))..... ( -30.70) >DroEre_CAF1 104032 94 + 1 GGCAGAGCAGUACUGAGUUUUGCCCUGCACAUUUAGGGCGCUAAAACGAUGCUCCAAUAAGCCGCAUUUUGGCUUAUUGCUGCCUUGUUUUUUC (((((((((......(((...((((((......))))))))).......)))))((((((((((.....)))))))))).)))).......... ( -34.92) >DroYak_CAF1 102249 94 + 1 GGCAGAGUAAAAUUGAGUUUUGUCCUGCACAUUAAGGGCGCUAAAACGAUGCCCCAAUAAGCCGUACUUUGGCUCAUUGCUGCUUUGUUUUUUC .((((..((((((...))))))..))))..............(((((((.((..((((.(((((.....))))).))))..)).)))))))... ( -25.40) >DroAna_CAF1 121039 93 + 1 AACAGAACAGACAUGAAUUUUGAAUUGAAUUCUAAAGGAGCUAGAACGACGCCACAAUCGGCUUAA-AUUAACUGAUUGUGGCCUUGUUUUUUC ..............((((((......))))))..........(((((((.(((((((((((.....-.....))))))))))).)))))))... ( -25.20) >consensus GGCAGAGCAGAAUUGAGUUUUGUCUUGCACAUUAAGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUUC .((((..((((((...))))))..))))..............(((((((.((..((((.(((((.....))))).))))..)).)))))))... (-19.76 = -19.68 + -0.08)


| Location | 5,227,389 – 5,227,483 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
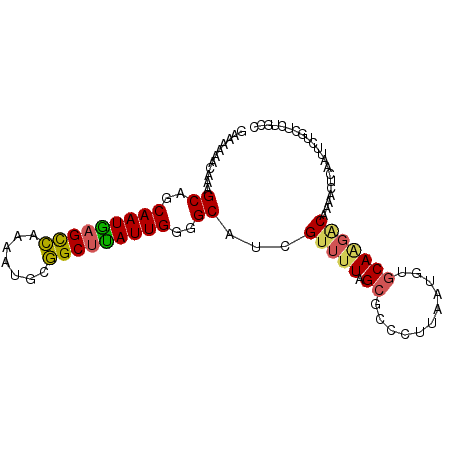
>2R_DroMel_CAF1 5227389 94 - 20766785 GCAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCGCCCUUAAUAUGCAAGACAAAACUCAAUUCUACUCUCCC ...........(((.((((((((((.....))))))))))((((..(.....)..))))......))).......................... ( -25.50) >DroSec_CAF1 103132 94 - 1 GCAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCUCCCUUAAUGUGCAAGACAAAACUCAAUUCUGCUCUGGC .......((.(((((((((((((((.....))))))))))..(((.((((..((...))..)))))))................))))).)).. ( -29.20) >DroSim_CAF1 89293 94 - 1 GUAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCUCCCGUAAUGUGCAAGACAAAACUCAAUUCUGCUCUGCC .......((.(((((((((((((((.....))))))))))..(((.((((.(((...))).)))))))................))))).)).. ( -28.30) >DroEre_CAF1 104032 94 - 1 GAAAAAACAAGGCAGCAAUAAGCCAAAAUGCGGCUUAUUGGAGCAUCGUUUUAGCGCCCUAAAUGUGCAGGGCAAAACUCAGUACUGCUCUGCC ..........((((.(((((((((.......)))))))))(((((..((....))(((((........)))))............))))))))) ( -32.10) >DroYak_CAF1 102249 94 - 1 GAAAAAACAAAGCAGCAAUGAGCCAAAGUACGGCUUAUUGGGGCAUCGUUUUAGCGCCCUUAAUGUGCAGGACAAAACUCAAUUUUACUCUGCC ...........(((((((((((((.......)))))))))(((....(((((.((((.......)))))))))....))).........)))). ( -24.90) >DroAna_CAF1 121039 93 - 1 GAAAAAACAAGGCCACAAUCAGUUAAU-UUAAGCCGAUUGUGGCGUCGUUCUAGCUCCUUUAGAAUUCAAUUCAAAAUUCAUGUCUGUUCUGUU (((...(((..(((((((((.(((...-...))).)))))))))...(((((((.....)))))))...............)))...))).... ( -22.70) >consensus GAAAAAACAAAGCAGCAAUGAGCCAAAAUGCGGCUUAUUGGGGCAUCGUUUUAGCGCCCUUAAUGUGCAAGACAAAACUCAAUUCUGCUCUGCC ...........((..(((((((((.......)))))))))..))...(((((.((...........)))))))..................... (-16.61 = -16.37 + -0.25)

| Location | 5,227,423 – 5,227,515 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -18.25 |
| Energy contribution | -17.92 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
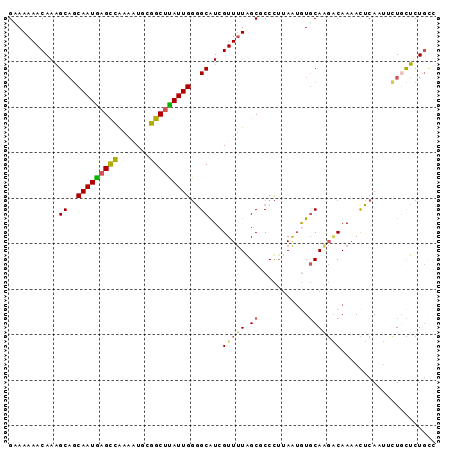
>2R_DroMel_CAF1 5227423 92 + 20766785 AGGGCGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUGCGACCUACUAACAUUGU--AACCAAUUCGACCAGU .((((((.(((((((.((..((((.(((((.....))))).))))..)).))))))).))).)))...........--................ ( -27.40) >DroSec_CAF1 103166 92 + 1 AGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUGCGACCUACUAACAUUGU--AAUCUAUUCGACCAGU (((..((.(((((((.((..((((.(((((.....))))).))))..)).))))))).))..)))...........--................ ( -26.80) >DroSim_CAF1 89327 92 + 1 CGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUACGACCUACUAACAUUGU--AAUCUAUUCGACCAGU .((..(..(((((((.((..((((.(((((.....))))).))))..)).)))))))..)..))............--................ ( -21.80) >DroEre_CAF1 104066 92 + 1 AGGGCGCUAAAACGAUGCUCCAAUAAGCCGCAUUUUGGCUUAUUGCUGCCUUGUUUUUUCGACCCACUAACAUUGU--AACCAAUUCGACCAGU .(((((..(((((((.((..((((((((((.....))))))))))..)).)))))))..)).)))...........--................ ( -30.10) >DroYak_CAF1 102283 92 + 1 AGGGCGCUAAAACGAUGCCCCAAUAAGCCGUACUUUGGCUCAUUGCUGCUUUGUUUUUUCGACCUACUAAAAUUGU--ACCAAAUUCGACCAGU .(((((..(((((((.((..((((.(((((.....))))).))))..)).)))))))..)).)))...........--................ ( -23.60) >DroAna_CAF1 121073 93 + 1 AAGGAGCUAGAACGACGCCACAAUCGGCUUAA-AUUAACUGAUUGUGGCCUUGUUUUUUCGUCCACCUAAAAUUAAUUCCUUAAUUCGACCAGU ..((((..(((((((.(((((((((((.....-.....))))))))))).)))))))..).))).............................. ( -27.00) >consensus AGGGAGCCAAAACGAUGCCCCAAUAAGCCGCAUUUCGGCUCAUUGCUGCUUUGUUUUUUCGACCUACUAACAUUGU__AACCAAUUCGACCAGU .(((....(((((((.((..((((.(((((.....))))).))))..)).))))))).....)))............................. (-18.25 = -17.92 + -0.33)



| Location | 5,227,423 – 5,227,515 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -18.29 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5227423 92 - 20766785 ACUGGUCGAAUUGGUU--ACAAUGUUAGUAGGUCGCAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCGCCCU (((((....((((...--.)))).)))))(((.(((.(((((...((..((((((((((.....))))))))))..))...))))).))).))) ( -32.30) >DroSec_CAF1 103166 92 - 1 ACUGGUCGAAUAGAUU--ACAAUGUUAGUAGGUCGCAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCUCCCU ..(((((.....))))--)..........(((..((.(((((...((..((((((((((.....))))))))))..))...))))).))..))) ( -28.80) >DroSim_CAF1 89327 92 - 1 ACUGGUCGAAUAGAUU--ACAAUGUUAGUAGGUCGUAAAAACAAAGCAGCAAUGAGCCGAAAUGCGGCUUAUUGGGGCAUCGUUUUGGCUCCCG ..(((((.....))))--)...........((..((.(((((...((..((((((((((.....))))))))))..))...))))).))..)). ( -27.40) >DroEre_CAF1 104066 92 - 1 ACUGGUCGAAUUGGUU--ACAAUGUUAGUGGGUCGAAAAAACAAGGCAGCAAUAAGCCAAAAUGCGGCUUAUUGGAGCAUCGUUUUAGCGCCCU (((((....((((...--.)))).)))))(((.((..(((((...((..(((((((((.......)))))))))..))...)))))..))))). ( -29.50) >DroYak_CAF1 102283 92 - 1 ACUGGUCGAAUUUGGU--ACAAUUUUAGUAGGUCGAAAAAACAAAGCAGCAAUGAGCCAAAGUACGGCUUAUUGGGGCAUCGUUUUAGCGCCCU ((..(......)..))--...........(((.((..(((((...((..(((((((((.......)))))))))..))...)))))..)).))) ( -25.10) >DroAna_CAF1 121073 93 - 1 ACUGGUCGAAUUAAGGAAUUAAUUUUAGGUGGACGAAAAAACAAGGCCACAAUCAGUUAAU-UUAAGCCGAUUGUGGCGUCGUUCUAGCUCCUU (((....(((((((....)))))))..)))(((.(....(((.(.(((((((((.(((...-...))).))))))))).).)))....)))).. ( -24.70) >consensus ACUGGUCGAAUUGAUU__ACAAUGUUAGUAGGUCGAAAAAACAAAGCAGCAAUGAGCCAAAAUGCGGCUUAUUGGGGCAUCGUUUUAGCGCCCU ..............................((..(..(((((...((..(((((((((.......)))))))))..))...)))))..)..)). (-18.29 = -17.68 + -0.61)

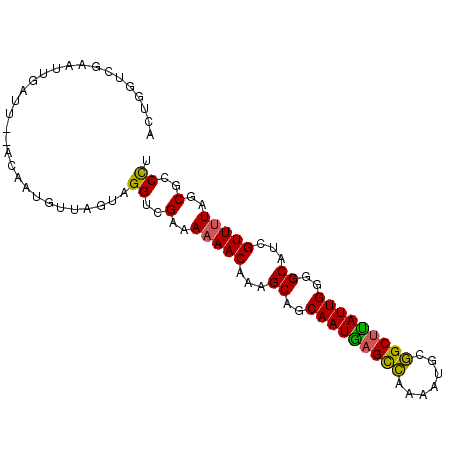
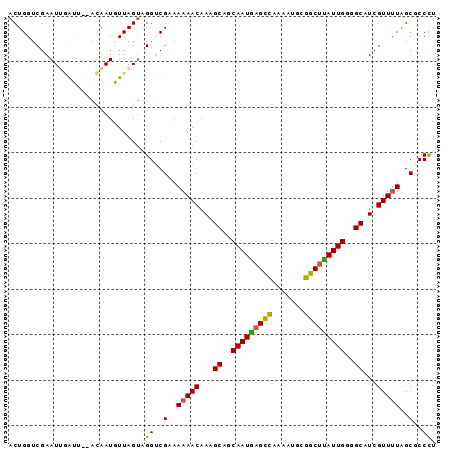
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:54 2006