
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,201,643 – 5,201,749 |
| Length | 106 |
| Max. P | 0.640533 |

| Location | 5,201,643 – 5,201,749 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.39 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
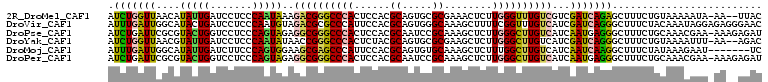
>2R_DroMel_CAF1 5201643 106 + 20766785 AUCUGGUUAACAUAUUGAUCCUCCCAAUAAAGACGGGCCCACUCCACGCAGUGCGCGAAACUCUUGGGUUUGUCGUCGAUCAGAGCUUUCUGUAAAAAUA-AA--UUAC .((((((..((.(((((.......)))))..((((((((((.....(((.....))).......))))))))))))..))))))................-..--.... ( -29.20) >DroVir_CAF1 107465 109 + 1 AUUUGAUUGGCAUACUGAUCCUCCCAAUGUAGACGCGCCCAUUCCACGCAGUGGGCAAAGCUUUUCGGUUUGUCAUCGAUCAGGGCUUUCUACAAAUAGGAGAGGGAAC ....(((..(....)..)))((((...((((((...(((((((......))))))).((((((((((((.....)))))..)))))))))))))....))))....... ( -33.50) >DroPse_CAF1 78524 108 + 1 AUCUGAUUCGCGUACUGGUCCUCCCAGUAGAGGCGGGCCCACUCCACGCAAUCCGCAAAGCUCUUGGGCUUGUCAUCAAUGAGGGCUUUCUGCAAACGAA-AAAGAGAU .(((..((((.(((..(((((((...((.((((((((((((......((.....))........)))))))))).)).)))))))))...)))...))))-..)))... ( -32.94) >DroYak_CAF1 77933 106 + 1 AUCUGGUUAACGUAUUGAUCCUCCCAAUAUAACCGGGCCCACUCUACGCAGUGCGCGAAGCUCUUGGGCUUGUCAUCGAUCAGGGCUUUCUGUAAAAUUU-AA--AGAC ..((((((...((((((.......))))))))))))((((......(((.....)))(((((....)))))...........))))..............-..--.... ( -26.50) >DroMoj_CAF1 113395 102 + 1 AUUUGAUUGGCAUAUUGAUCUUCCCAGUGGAAGCGAGCCCAUUCCACGCAGUGUGCAAAGCUCUUUGGCUUGUCAUCAAUCAAGGCUUUCUAUAAAGAAU-------UC .((((((((((((((((.........((((((.........)))))).)))))))).(((((....))))).....))))))))...((((....)))).-------.. ( -27.50) >DroPer_CAF1 81266 108 + 1 AUCUGAUUCGCGUACUGGUCCUCCCAGUAGAGGCGGGCCCACUCCACGCAAUCCGCAAAGCUCUUGGGCUUGUCAUCAAUGAGGGCUUUCUGCAAACGAA-AAAGAGAU .(((..((((.(((..(((((((...((.((((((((((((......((.....))........)))))))))).)).)))))))))...)))...))))-..)))... ( -32.94) >consensus AUCUGAUUAGCAUACUGAUCCUCCCAAUAGAGGCGGGCCCACUCCACGCAGUGCGCAAAGCUCUUGGGCUUGUCAUCAAUCAGGGCUUUCUGCAAAAAAA_AA__AGAC .(((((((....(((((.......)))))..((((((((((......((.....))........))))))))))...)))))))......................... (-19.91 = -19.39 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:45 2006