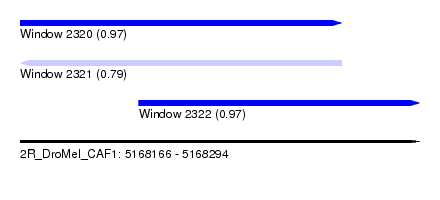
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,168,166 – 5,168,294 |
| Length | 128 |
| Max. P | 0.974650 |

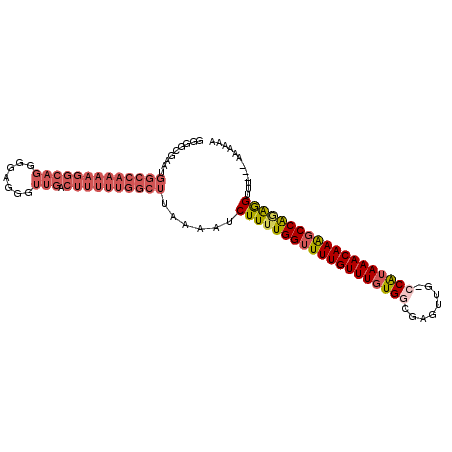
| Location | 5,168,166 – 5,168,269 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -28.05 |
| Energy contribution | -30.17 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5168166 103 + 20766785 GGGGCGAAUGGCCAAAAGGCAGGGGAGGGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG-CCAUAAACAAAGCCAGAGGUUU---AAAAAG .........(((((((((((((.......))).))))))))))..((((((((.(((((((((((((((.....)-))))))))))))))))))))))---...... ( -41.30) >DroSec_CAF1 43798 103 + 1 GGGGCGAAUGGCCAAAAGGCAGGGGUGAGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG-GCAUAAACAAAGCCAGAGGUUU---AAACAG .........(((((((((((((.......))).))))))))))..((((((((.(((((((((((((.(.....)-.)))))))))))))))))))))---...... ( -36.70) >DroSim_CAF1 30352 104 + 1 GGGGCGAAUGGCCAAAAGGCAGGGGAAGGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUGGGCAAAAACAAACCCAGAAGUUU---AAAAAA .........(((((((((((((.......))).))))))))))......(((((((.(((((((.((.(......).)).))))))).)))))))...---...... ( -29.30) >DroEre_CAF1 45751 106 + 1 GGGGCGAAUGGCCAAAAGGCAGGGGAGGGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAAUUG-CCAUAAACAAAGCCGGAGGUUUUCUAAGGAA .....(((.(((((((((((((.......))).))))))))))......((((((((((((((((((((.....)-)))))))))))))))))))..)))....... ( -40.90) >DroYak_CAF1 44050 102 + 1 GGGGCGAAUGGCCAAAAGGCAGGGGAGGGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGG-GAGUUG-CCAUAAACAAAGCCGAAGGUUU---AAAAAA .........(((((((((((((.......))).))))))))))..((((((((.((((((((((((((-......-))))))))))))))))))))))---...... ( -38.00) >DroAna_CAF1 65743 79 + 1 -----------------------GCAGGGUUGACUUUUUGGCUUAAAAUCUU-UGGUUUUGUUUGUGGCCAGUUG-CCGUAAACAAGGCCAGGGGAUU---UAAUAA -----------------------....(((..(....)..)))((((.((((-((((((((((((((((.....)-))))))))))))))))))).))---)).... ( -27.70) >consensus GGGGCGAAUGGCCAAAAGGCAGGGGAGGGUUGACUUUUUGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG_CCAUAAACAAAGCCAGAGGUUU___AAAAAA .........(((((((((((((.......))).))))))))))......(((((((((((((((((((........)))))))))))))))))))............ (-28.05 = -30.17 + 2.12)

| Location | 5,168,166 – 5,168,269 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -18.85 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5168166 103 - 20766785 CUUUUU---AAACCUCUGGCUUUGUUUAUGG-CAACUCGCCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACCCUCCCCUGCCUUUUGGCCAUUCGCCCC ......---........((((((((((.(((-(.....)))).)))))))...............(((((((.((............)).))))))).....))).. ( -22.90) >DroSec_CAF1 43798 103 - 1 CUGUUU---AAACCUCUGGCUUUGUUUAUGC-CAACUCGCCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACUCACCCCUGCCUUUUGGCCAUUCGCCCC ......---........((((((((((.((.-(.....).)).)))))))...............(((((((.((............)).))))))).....))).. ( -15.80) >DroSim_CAF1 30352 104 - 1 UUUUUU---AAACUUCUGGGUUUGUUUUUGCCCAACUCGCCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACCUUCCCCUGCCUUUUGGCCAUUCGCCCC ......---...(((.(((.(((((((.((.(......).)).))))))).))).))).......(((((((.((............)).))))))).......... ( -17.80) >DroEre_CAF1 45751 106 - 1 UUCCUUAGAAAACCUCCGGCUUUGUUUAUGG-CAAUUCGCCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACCCUCCCCUGCCUUUUGGCCAUUCGCCCC .......(((.......((.(((((((.(((-(.....)))).))))))).))............(((((((.((............)).)))))))..)))..... ( -23.50) >DroYak_CAF1 44050 102 - 1 UUUUUU---AAACCUUCGGCUUUGUUUAUGG-CAACUC-CCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACCCUCCCCUGCCUUUUGGCCAUUCGCCCC ......---........((((((((((.(((-......-))).)))))))...............(((((((.((............)).))))))).....))).. ( -19.20) >DroAna_CAF1 65743 79 - 1 UUAUUA---AAUCCCCUGGCCUUGUUUACGG-CAACUGGCCACAAACAAAACCA-AAGAUUUUAAGCCAAAAAGUCAACCCUGC----------------------- ......---.......(((..((((((..((-(.....)))..))))))..)))-..((((((.......))))))........----------------------- ( -13.90) >consensus UUUUUU___AAACCUCUGGCUUUGUUUAUGG_CAACUCGCCACAAACAAAACCAAAAGAUUUUAAGCCAAAAAGUCAACCCUCCCCUGCCUUUUGGCCAUUCGCCCC .................((.(((((((.(((........))).))))))).))....((((((.......))))))...........(((....))).......... (-12.54 = -13.33 + 0.79)

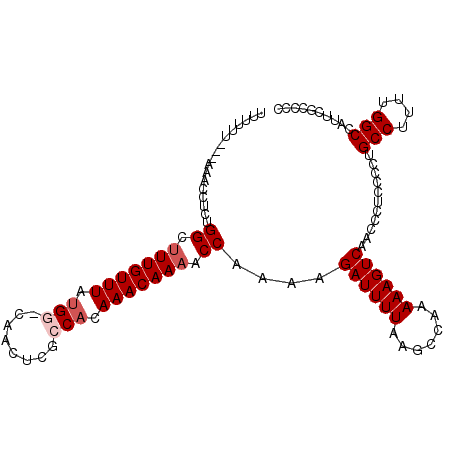
| Location | 5,168,204 – 5,168,294 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.32 |
| Mean z-score | -4.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
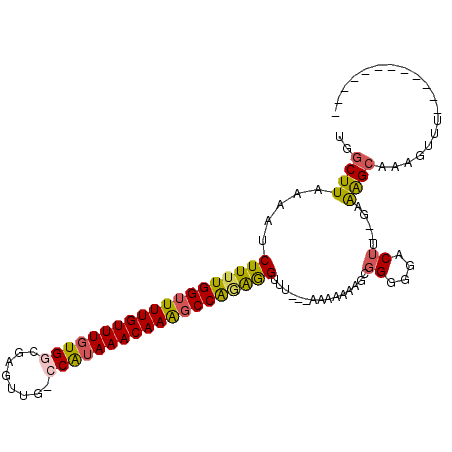
>2R_DroMel_CAF1 5168204 90 + 20766785 UGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG-CCAUAAACAAAGCCAGAGGUUU---AAAAAGAGCGGGGGACUU--GAAAGCAAAGUUU------------ ..((((.....((((((((((((((((((((.....)-)))))))))))))))))))...---.....))))...((((((--.......))))))------------ ( -34.32) >DroSec_CAF1 43836 90 + 1 UGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG-GCAUAAACAAAGCCAGAGGUUU---AAACAGAGCGGGGGACUU--GAAAGCAAAGUUU------------ ..((((.....((((((((((((((((((.(.....)-.))))))))))))))))))...---.....))))...((((((--.......))))))------------ ( -29.72) >DroSim_CAF1 30390 91 + 1 UGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUGGGCAAAAACAAACCCAGAAGUUU---AAAAAAAACGGGGGACUU--GAAAGCAAAGUUU------------ ..((((.....(((((((.(((((((.((.(......).)).))))))).)))))))...---.........((....)).--..)))).......------------ ( -22.30) >DroEre_CAF1 45789 91 + 1 UGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAAUUG-CCAUAAACAAAGCCGGAGGUUUUCUAAGGAA--CGGGGGACUU--GAAAGCAAAGUUU------------ ..((((........(((((((((((((((((.....)-))))))))))))))))(((((..((......--.))..)))))--..)))).......------------ ( -34.20) >DroYak_CAF1 44088 89 + 1 UGGCUUAAAAUCUUUUGGUUUUGUUUGUGG-GAGUUG-CCAUAAACAAAGCCGAAGGUUU---AAAAAAUGUGGGGGACUG--GAAAGCAAAGUUU------------ ..((((.((((((((.((((((((((((((-......-))))))))))))))))))))))---......(.(((....)))--.))))).......------------ ( -31.80) >DroAna_CAF1 65758 103 + 1 UGGCUUAAAAUCUU-UGGUUUUGUUUGUGGCCAGUUG-CCGUAAACAAGGCCAGGGGAUU---UAAUAAGGACGUGGACUACUAAGGGAAAAGUUUCCCACUGCCAGG (((((((((.((((-((((((((((((((((.....)-))))))))))))))))))).))---)))...................(((((....)))))...)))).. ( -38.40) >consensus UGGCUUAAAAUCUUUUGGUUUUGUUUGUGGCGAGUUG_CCAUAAACAAAGCCAGAGGUUU___AAAAAAAGCGGGGGACUU__GAAAGCAAAGUUU____________ ..((((.....(((((((((((((((((((........)))))))))))))))))))...............((....)).....))))................... (-25.08 = -25.08 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:39 2006