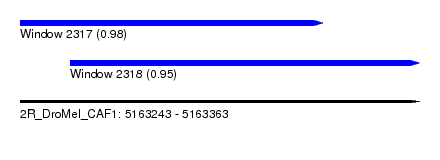
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,163,243 – 5,163,363 |
| Length | 120 |
| Max. P | 0.982239 |

| Location | 5,163,243 – 5,163,334 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
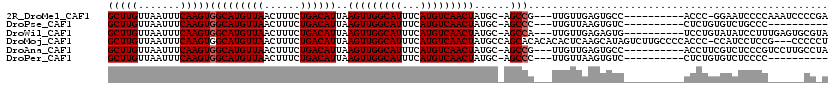
>2R_DroMel_CAF1 5163243 91 + 20766785 ----------GAAGGAC---------UCU----GCGCGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCG---UUGUUG ----------.((.(((---------.((----((((.(((((.......))))).))((((((......))))))..(((((((((...)))))))))..))-))..)---)).)). ( -28.60) >DroVir_CAF1 52191 95 + 1 ----------GU--UCG---------CUUUGGUUAGGGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGCCAGCGC--ACACUCA ----------((--.((---------((..........(((((.......)))))(((((((((......))))....(((((((((...)))))))))))))))))).--))..... ( -27.30) >DroPse_CAF1 39069 110 + 1 GUGUGUGUGUGGAGGACUGCACCGUGUGU----GAGCGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCC---UUGUUA ...........((((.(((((...(((((----((((.(((((.......))))).))((((((((((.........)))))))))).))))).))....)))-)).))---)).... ( -33.00) >DroWil_CAF1 50128 91 + 1 ----------UAAGUGU---------GUG----GCGCGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCA---UUGUUG ----------.......---------(((----((((.(((((.......))))).))((((((......))))))..(((((((((...)))))))))....-.))))---)..... ( -28.00) >DroMoj_CAF1 53897 98 + 1 ----------GCGCCAG---------CCUGGG-CUGGGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGCCAGCACACACACUCA ----------((.((((---------(....)-)))).(((((.......)))))(((((((((......))))....(((((((((...)))))))))))))).))........... ( -33.00) >DroPer_CAF1 38860 110 + 1 GUGUGUGUGUGGAGGACUGCACCGUGUGU----GAGCGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCC---UUGUUA ...........((((.(((((...(((((----((((.(((((.......))))).))((((((((((.........)))))))))).))))).))....)))-)).))---)).... ( -33.00) >consensus __________GAAGGAC_________UCU____GAGCGGCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC_AGCCC___UUGUUA ...................................((.(((((.......))))).))((((((......))))))..(((((((((...)))))))))................... (-18.63 = -18.97 + 0.33)

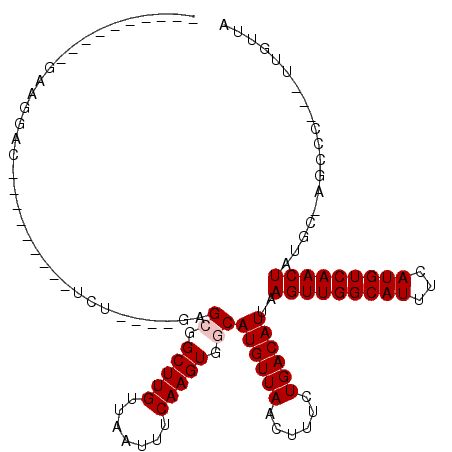
| Location | 5,163,258 – 5,163,363 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5163258 105 + 20766785 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCG---UUGUUGAGUGCC----------ACCC-GGAAUCCCCAAAUCCCCGA ..(((...(((((..(((((((.(((((...(((.(((..(((((((((...)))))))))))))-))...---..))))))))))----------))..-)))))...)))........ ( -29.60) >DroPse_CAF1 39103 96 + 1 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCC---UUGUUAAGUGUC----------CUCUGUGUCUGCCC---------- (((((.......)))))(((((.(((((...(((.(((..(((((((((...)))))))))))))-))...---..))))))))))----------..............---------- ( -22.70) >DroWil_CAF1 50143 106 + 1 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCA---UUGUUGAGAGUG----------UCCUGUAUAUCCUUUGAGUGCGUA (((((.......)))))(((((.(((((...(((.(((..(((((((((...)))))))))))))-))...---..)))))..)))----------))..((((.((....))))))... ( -24.70) >DroMoj_CAF1 53915 116 + 1 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGCCAGCACACACACUCAAGCAUAGUCUUGCCCCACCC-CCAUCCUCCG---CCCCCU (((((...........((((((((((......))))....(((((((((...)))))))))))))))...........))))).................-..........---...... ( -24.75) >DroAna_CAF1 60836 106 + 1 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCG---UUGUUGAGUGCC----------ACCUUCGUCUCCCGUCCUUGCCUA ...............(((((((.(((((...(((.(((..(((((((((...)))))))))))))-))...---..))))))))))----------))...................... ( -26.60) >DroPer_CAF1 38894 96 + 1 GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC-AGCCC---UUGUUAAGUGUC----------CUCUGUGUCUCCCC---------- (((((.......)))))(((((.(((((...(((.(((..(((((((((...)))))))))))))-))...---..))))))))))----------..............---------- ( -22.70) >consensus GCUUGUUAAUUUCAAGUGGCAUGUUAACUUUCUGACAUUAAGUUGGCAUUUCAUGUCAACUAUGC_AGCCC___UUGUUAAGUGUC__________ACCUGUGUCUCCCC______CC_A (((((.......)))))(((((((((......))))))..(((((((((...)))))))))......))).................................................. (-19.22 = -19.38 + 0.17)
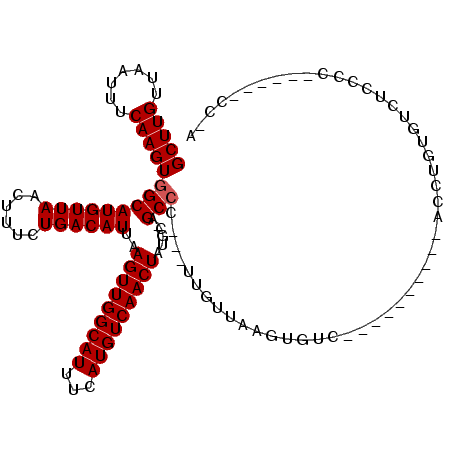
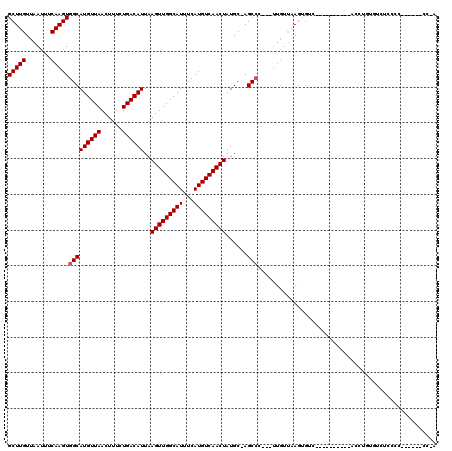
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:35 2006