
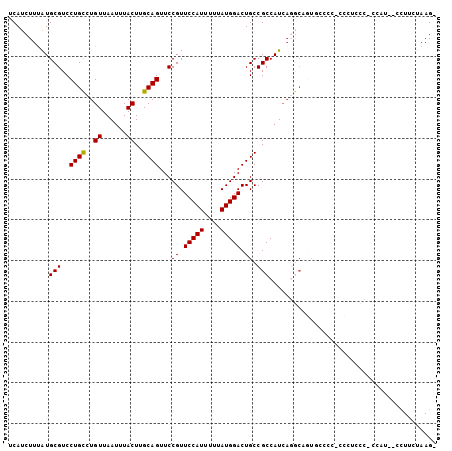
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,144,195 – 5,144,314 |
| Length | 119 |
| Max. P | 0.912860 |

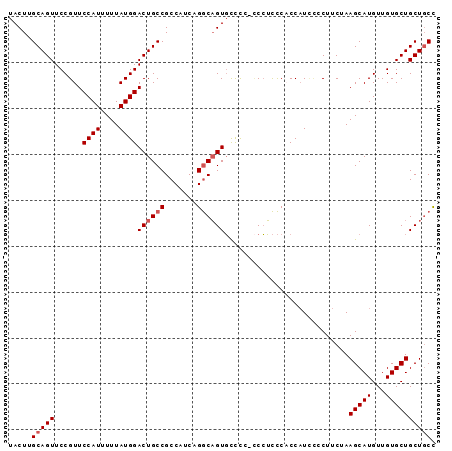
| Location | 5,144,195 – 5,144,297 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -17.68 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5144195 102 + 20766785 UCAUCUUUGUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGCCU--CCCUCCCCCCCUUUCCUUCUAAG- ............(..(((((((...........(((((.((((.........)))))))))......)))))))..)..--.......................- ( -22.13) >DroSec_CAF1 20058 103 + 1 UCAUCUUUAUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGUUCC-CCCUCCCACCAUCCCCUUCUAAG- ........(((.(..((((..((......))..))))..)))).((((....))))((((((.......)))))).....-.......................- ( -20.40) >DroSim_CAF1 6171 104 + 1 UCAUCUUUAUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGUCCCAACCUCCCCCCAUCCCCUUCUAAG- .........((.(.((((((((...........(((((.((((.........)))))))))......))))))).).).)).......................- ( -21.53) >DroEre_CAF1 22018 84 + 1 UCAUCUUUAUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAU--------UCCCC-UCC-----------CCUUCUAAG- ..........(((..((((..((......))..))))...((.(((((....))))).)).)))...--------.....-...-----------.........- ( -13.00) >DroYak_CAF1 20147 101 + 1 UCAUCUUUAUGCGUCCUGUCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGACUGUUCCCC-UCAGUUCACCAU--GCUUCUAAG- ..........((((..((...............(((((.((((.........)))))))))........(((((......-.)))))))..))--)).......- ( -16.00) >DroAna_CAF1 42105 89 + 1 UCAUCUUCAUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAUGCU----UCC-CCCU-----------UUUCUAAGC ..........(((..((((..((......))..))))...((.(((((....))))).)).))).........----...-....-----------......... ( -13.00) >consensus UCAUCUUUAUGCGUCCUGCCUGUUAAUUUACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGCCCC_CCCUCCC_CCAU__CCUUCUAAG_ ..........(((..((((..((......))..))))...((.(((((....))))).)).)))......................................... (-12.92 = -12.78 + -0.14)


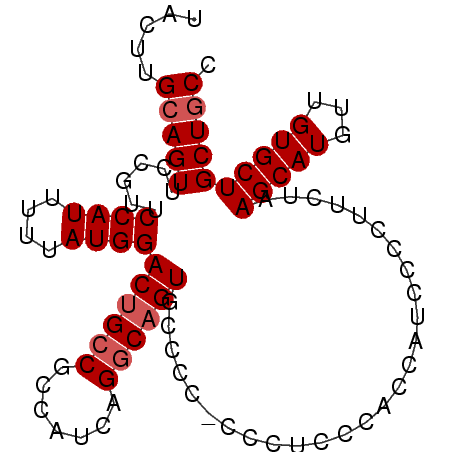
| Location | 5,144,223 – 5,144,314 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.75 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5144223 91 + 20766785 UACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGCCU--CCCUCCCCCCCUUUCCUUCUAAGCAUGUUGUGCUGCUGCC .....(((((......((((....))))((((((.......))))))....--.....................(((((...)))))))))). ( -21.90) >DroSec_CAF1 20086 92 + 1 UACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGUUCC-CCCUCCCACCAUCCCCUUCUAAGCAUGUUGUGCUGCUGCC .....(((((......((((....))))((((((.......)))))).....-.....................(((((...)))))))))). ( -21.90) >DroSim_CAF1 6199 93 + 1 UACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGUCCCAACCUCCCCCCAUCCCCUUCUAAGCAUGUUGUGCUGCUGCC .....(((((......((((....))))((((((.......))))))...........................(((((...)))))))))). ( -21.90) >DroYak_CAF1 20175 90 + 1 UACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGACUGUUCCCC-UCAGUUCACCAU--GCUUCUAAGCAUGUUGUGCUGCUCCU .....(((((.((((.........)))))))))...................-.((((.((.(((--(((....)))))).)).))))..... ( -22.30) >consensus UACUUGCAGUUCCGUUCCAUUUUUAUGGACUGCCGCCAUCAGGCAGUGCCCC_CCCUCCCACCAUCCCCUUCUAAGCAUGUUGUGCUGCUGCC .....(((((......((((....))))((((((.......))))))...........................(((((...)))))))))). (-17.00 = -17.75 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:24 2006