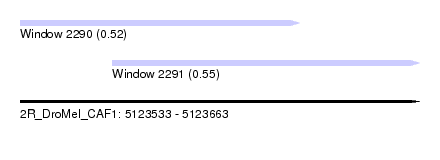
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,123,533 – 5,123,663 |
| Length | 130 |
| Max. P | 0.549302 |

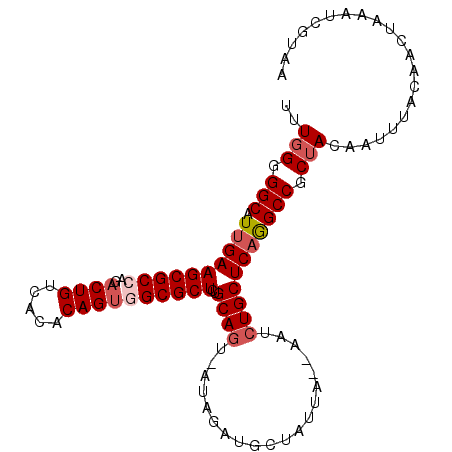
| Location | 5,123,533 – 5,123,624 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.33 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5123533 91 + 20766785 AAAUCUAAGGAAAGGCCGCUCG-GUGUCGUCUUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGCUGCGCUCUGCAGUUAAAAAUGC-AUUA- ...((((((((.(.(((....)-)).)..))))))))........(((((.((.(((.....)))))))))).((((........)))-)...- ( -25.70) >DroPse_CAF1 28517 79 + 1 UAAUUCUAUAAAAGGCCGCUCGGGUGUCGUCUUUGGGGGCAAUCAAGCGCCAGACUGUCACACAGUUGCGCUCUGCUCA--------------- ..........((((((.((......)).))))))..(((((....(((((..(((((.....)))))))))).))))).--------------- ( -24.50) >DroSec_CAF1 32745 92 + 1 AAACCUAAGGAAAGGCCGCUCG-GUGUCGUCUUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUGGCGCUCUGCAGU-AUAGAUGCUAUUAA ...((((((((.(.(((....)-)).)..))))))))((((((..((((((..((((.....)))))))))).......-...))))))..... ( -32.30) >DroSim_CAF1 52593 92 + 1 AAACCUAAGGAAAGGCCGCUCG-GUGUCGUCUUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUGGCGCUCUGCAGU-AUAGAUGCUAUUAA ...((((((((.(.(((....)-)).)..))))))))((((((..((((((..((((.....)))))))))).......-...))))))..... ( -32.30) >DroYak_CAF1 30154 91 + 1 AAAUCUAAGGAAAGGCCGCUCG-GUGUCGUCUUUUGGGGCAUGGAAGCGCCAGACUGUCACACAGUUGCGCUCUGCAUU-AUAGAUCCUAUUA- ..(((((.((.....))((((.-(((((.((....))))))).))(((((..(((((.....))))))))))..))...-.))))).......- ( -25.10) >DroPer_CAF1 27949 79 + 1 UAAUUCUAUAAAAGGCCGCUCGGGUGUCGUCUUUGGGGGCAAUCAAGCGCCAGACUGUCACACAGUUGCGCUCUGCUCA--------------- ..........((((((.((......)).))))))..(((((....(((((..(((((.....)))))))))).))))).--------------- ( -24.50) >consensus AAAUCUAAGGAAAGGCCGCUCG_GUGUCGUCUUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUUGCGCUCUGCAGU_AUAGAUGC_AUUA_ ...(((....((((((.((......)).)))))).)))(((....(((((..(((((.....)))))))))).))).................. (-20.50 = -20.33 + -0.16)

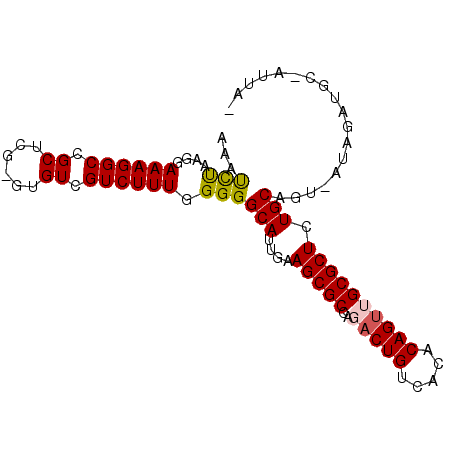
| Location | 5,123,563 – 5,123,663 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -21.81 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5123563 100 + 20766785 UUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGCUGCGCUCUGCAGUUAAAAAUGC-AUUA--AAUCUGCUCAGGCCGCUACAAUUUACAACUCAAUCUUAA ..(((.(((.((((.(((((((.(((.....)))))).))..((((........)))-)...--.....))))))))).)))..................... ( -25.90) >DroSec_CAF1 32775 102 + 1 UUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUGGCGCUCUGCAGU-AUAGAUGCUAUUAAAAAUCUGCUCAGGCCGCUACAAUUUACAACUAAAUCGUAA ..(((.(((.....((((((..((((.....))))))))))(((.(((-..((((..........))))))))))))).)))..................... ( -28.20) >DroSim_CAF1 52623 102 + 1 UUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUGGCGCUCUGCAGU-AUAGAUGCUAUUAAAAAUCUGCUCAGGCCGCUACAAUUUACAACUAAAUCGUAA ..(((.(((.....((((((..((((.....))))))))))(((.(((-..((((..........))))))))))))).)))..................... ( -28.20) >DroYak_CAF1 30184 100 + 1 UUUUGGGGCAUGGAAGCGCCAGACUGUCACACAGUUGCGCUCUGCAUU-AUAGAUCCUAUUA--AAUCUGCUCAAGCCGCUAGAAUUUACAGCAAAAUCUUGA ((((((((((((.((((((..(((((.....)))))))))).).))).-.(((((.......--.))))).....))).)))))).................. ( -24.20) >consensus UUUGGGGGCAUUGAAGCGCCAGACUGUCACACAGUGGCGCUCUGCAGU_AUAGAUGCUAUUA__AAUCUGCUCAGGCCGCUACAAUUUACAACUAAAUCGUAA ..(((.(((.((((((((((..((((.....))))))))))..((((....................))))))))))).)))..................... (-21.81 = -23.12 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:09 2006