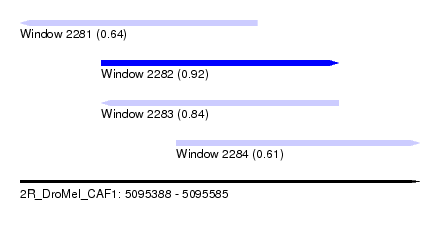
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,095,388 – 5,095,585 |
| Length | 197 |
| Max. P | 0.924378 |

| Location | 5,095,388 – 5,095,505 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5095388 117 - 20766785 AGAGCUUUGAGCAGCAGCUGGAAGACCUGAAGCAGAAACUGGAGCUGCUAAA---GAACCAACCCAGAGAGCAGGAAACACAAGGGAAAGUGGUGGAUAAAUCUCUAGGGGAAAUGUAAG ....((((.(((....))).))))..((..(((((.........)))))..)---)......((((((((........(((........))).........))))).))).......... ( -27.63) >DroSec_CAF1 1804 117 - 1 AGAACUUCAAGCAGCAGCUGGAAGAACUGAAGCAGAAACUGGAGCUGCUGAA---GAACCAACCCAAAGAGCAGGAAACGCUAGCGAAAGUGGUGGAUAAAUCUCUAGGGAAAAUGUAAG ....(((((.(((((..(..(.....(((...)))...)..).)))))))))---)......(((..((((.......(((((.(....)))))).......)))).))).......... ( -30.54) >DroSim_CAF1 1763 117 - 1 AGAACUUUCAGCAGCAGCUGGAAGACCUGAAGCAGAAACUGGAGCUGCUAAA---GAACCAACCCAAAGAGCAGGAAACGCAAGCGAAAGUGGUGGAUAAAUCUCUAGGGGAAAUGUAAG ....((((((((....))))))))..((..(((((.........)))))..)---)......(((..((((.......(((..((....)).))).......))))..)))......... ( -32.84) >DroYak_CAF1 1769 117 - 1 AGAGCUUUCAGCAGCAGCUGGAGAAGCUGAAGCAGAAACUGGAUCUACUAAA---GAACCAACCCAAAGAGCAGGAAACUCAAGAGAAAGUGGAGGAGAAGGCUUUAGGGGAAAUGUAAG ..((.((((.((..(((((.....)))))..)).))))))((.(((.....)---)).))..(((..(((((......(((...........)))......)))))..)))......... ( -29.40) >DroAna_CAF1 1630 120 - 1 AAUCUUUUCUUCAGCAAUUGGAAAACUUAAAGAACGAAUUGGAGAUACUGAAACGGAAGGAAGCGGAUGCCACCGCAUCGCAACAAAAACAAGAUAUUAAAAGCUUAGAAAAAAUGUAUG ..(((((((((((((((((.(.............).)))))......)))))..))))))).(((.((((....)))))))....................................... ( -17.72) >consensus AGAACUUUCAGCAGCAGCUGGAAGACCUGAAGCAGAAACUGGAGCUGCUAAA___GAACCAACCCAAAGAGCAGGAAACGCAAGAGAAAGUGGUGGAUAAAUCUCUAGGGGAAAUGUAAG ....((((((((....))))))))......(((((.........))))).............(((..((((.......(((...........))).......))))..)))......... (-11.98 = -13.94 + 1.96)


| Location | 5,095,428 – 5,095,545 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.45 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5095428 117 + 20766785 UGUUUCCUGCUCUCUGGGUUGGUUC---UUUAGCAGCUCCAGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUCAAAGCUCUCUUUAAGCUGCAGCUGCCCGAGAUAAUCCAAAUGCUCCUC ........((....((((((....(---(..(((((.........)))))..))(((((.(((((((.(((.((((....)))).))).)))))))...)))))))))))...))..... ( -34.10) >DroSec_CAF1 1844 117 + 1 CGUUUCCUGCUCUUUGGGUUGGUUC---UUCAGCAGCUCCAGUUUCUGCUUCAGUUCUUCCAGCUGCUGCUUGAAGUUCUCCUUAAGCUGCAGCUGCCCGAGAUGUUCCAAAUGCUCCUC ........((..((((((.....((---((..((((((.((((((..(....((..((((.(((....))).))))..)).)..)))))).))))))..))))...)))))).))..... ( -35.50) >DroSim_CAF1 1803 117 + 1 CGUUUCCUGCUCUUUGGGUUGGUUC---UUUAGCAGCUCCAGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUGAAAGUUCUCCUUAAGCUGCAGCUGCCCGAGAUGUUCCAAAUGCUCCUC ........((..((((((.....((---((..((((((.((((((..(....((..(((.((((....)))).)))..)).)..)))))).))))))..))))...)))))).))..... ( -35.20) >DroEre_CAF1 1820 117 + 1 UGUUUCCUGCUCGUUGGCUUGGUUC---UUGAGUAGCUCCAGUUUCUGCUUCAGGUUGUCCAGCUGCUGUUGGAAGCUCUCGUUAAGCUUCAGCUUCUCCAGACAAUCCAAGUGCUCCUC ................((((((.((---(.(((.((((..(((((..((....((((.((((((....))))))))))...)).)))))..)))).))).)))....))))))....... ( -31.80) >DroYak_CAF1 1809 117 + 1 AGUUUCCUGCUCUUUGGGUUGGUUC---UUUAGUAGAUCCAGUUUCUGCUUCAGCUUCUCCAGCUGCUGCUGAAAGCUCUCGUUGAGCUGCAGCUGCUCUAGACAAUCCAAGUGUUCCUC ........((.(((.((((((...(---(..((((((.......))))))..))..(((..(((.(((((....(((((.....)))))))))).)))..)))))))))))).))..... ( -36.90) >DroAna_CAF1 1670 120 + 1 CGAUGCGGUGGCAUCCGCUUCCUUCCGUUUCAGUAUCUCCAAUUCGUUCUUUAAGUUUUCCAAUUGCUGAAGAAAAGAUUCAUUUAAUUGCAUCUGCUCCAAGCUGGAGAGAUGCUUUUC .((.((((.(((....))).....)))).)).........((((..(((((((.((.........)))))))))..)))).........((((((.((((.....))))))))))..... ( -29.80) >consensus CGUUUCCUGCUCUUUGGGUUGGUUC___UUUAGCAGCUCCAGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUGAAAGCUCUCCUUAAGCUGCAGCUGCCCGAGACAAUCCAAAUGCUCCUC ....(((........)))((((..........((((((.((((((.......((..(((.((((....)))).)))..))....)))))).))))))..........))))......... (-18.64 = -18.45 + -0.19)

| Location | 5,095,428 – 5,095,545 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5095428 117 - 20766785 GAGGAGCAUUUGGAUUAUCUCGGGCAGCUGCAGCUUAAAGAGAGCUUUGAGCAGCAGCUGGAAGACCUGAAGCAGAAACUGGAGCUGCUAAA---GAACCAACCCAGAGAGCAGGAAACA .....((.(((((......((((((((((((.((((((((....)))))))).))))))).....)))))(((((.........)))))...---........)))))..)).(....). ( -41.50) >DroSec_CAF1 1844 117 - 1 GAGGAGCAUUUGGAACAUCUCGGGCAGCUGCAGCUUAAGGAGAACUUCAAGCAGCAGCUGGAAGAACUGAAGCAGAAACUGGAGCUGCUGAA---GAACCAACCCAAAGAGCAGGAAACG .....((.(((((......((...(((((((.(((((((.....))).)))).)))))))...)).((..(((((.........)))))..)---).......)))))..)).(....). ( -36.50) >DroSim_CAF1 1803 117 - 1 GAGGAGCAUUUGGAACAUCUCGGGCAGCUGCAGCUUAAGGAGAACUUUCAGCAGCAGCUGGAAGACCUGAAGCAGAAACUGGAGCUGCUAAA---GAACCAACCCAAAGAGCAGGAAACG .....((.(((((....(((..(((((((.((((((.(((....((((((((....)))))))).))).))))......)).)))))))..)---))......)))))..)).(....). ( -40.90) >DroEre_CAF1 1820 117 - 1 GAGGAGCACUUGGAUUGUCUGGAGAAGCUGAAGCUUAACGAGAGCUUCCAACAGCAGCUGGACAACCUGAAGCAGAAACUGGAGCUACUCAA---GAACCAAGCCAACGAGCAGGAAACA .....((.(((((.((((((.(((.(((((((((((.....)))))))...(((...(((............)))...))).)))).))).)---))..))).)))).).)).(....). ( -32.10) >DroYak_CAF1 1809 117 - 1 GAGGAACACUUGGAUUGUCUAGAGCAGCUGCAGCUCAACGAGAGCUUUCAGCAGCAGCUGGAGAAGCUGAAGCAGAAACUGGAUCUACUAAA---GAACCAACCCAAAGAGCAGGAAACU .........((((....(((..(((.((((((((((.....)))))....))))).)))..))).((....))......(((.(((.....)---)).)))..)))).....((....)) ( -31.40) >DroAna_CAF1 1670 120 - 1 GAAAAGCAUCUCUCCAGCUUGGAGCAGAUGCAAUUAAAUGAAUCUUUUCUUCAGCAAUUGGAAAACUUAAAGAACGAAUUGGAGAUACUGAAACGGAAGGAAGCGGAUGCCACCGCAUCG .....((((((((((.....)))).))))))........((.(((((((((((((((((.(.............).)))))......)))))..))))))).((((......)))).)). ( -31.92) >consensus GAGGAGCAUUUGGAUUAUCUCGAGCAGCUGCAGCUUAACGAGAGCUUUCAGCAGCAGCUGGAAAACCUGAAGCAGAAACUGGAGCUACUAAA___GAACCAACCCAAAGAGCAGGAAACG .........((((.........((.((((.(((...........((((((((....))))))))..(((...)))...))).)))).)).........)))).((........))..... (-13.96 = -14.77 + 0.81)


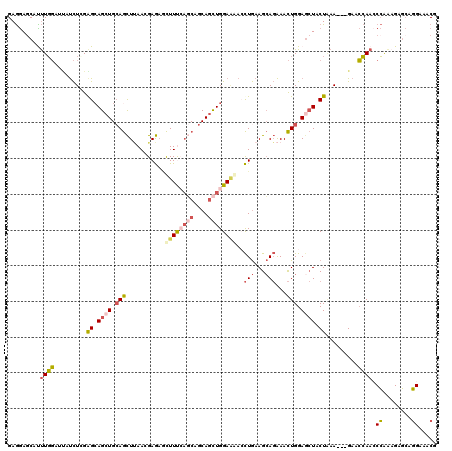
| Location | 5,095,465 – 5,095,585 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -19.77 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5095465 120 + 20766785 AGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUCAAAGCUCUCUUUAAGCUGCAGCUGCCCGAGAUAAUCCAAAUGCUCCUCUAGCGAAGCAAUCUUCUUCAGCUUCCAGGCGCAGAAGGCC ......((((((..(((((.(((((((.(((.((((....)))).))).)))))))...))))).........(((.....))))))))).((((((...(((....)))..)))))).. ( -40.00) >DroSec_CAF1 1881 120 + 1 AGUUUCUGCUUCAGUUCUUCCAGCUGCUGCUUGAAGUUCUCCUUAAGCUGCAGCUGCCCGAGAUGUUCCAAAUGCUCCUCCAGCGAAGCGAUCUUCUUCAGCUUCCAGGCGCAGUAGGCC .((((((((............((((((.((((((........)))))).))))))(((.(((..((.......))..)))..(.(((((((......)).)))))).))))))).)))). ( -38.90) >DroSim_CAF1 1840 120 + 1 AGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUGAAAGUUCUCCUUAAGCUGCAGCUGCCCGAGAUGUUCCAAAUGCUCCUCCAGCGAAGCGAUCUUUUUCAGCUUCCAGGCGCAGUAGGCC .((((((((.....(((((.(((((((.(((..(((.....))).))).)))))))...)))))...((....(((.....)))(((((((......)).)))))..)).)))).)))). ( -37.70) >DroEre_CAF1 1857 120 + 1 AGUUUCUGCUUCAGGUUGUCCAGCUGCUGUUGGAAGCUCUCGUUAAGCUUCAGCUUCUCCAGACAAUCCAAGUGCUCCUCCAGCAAAGCAAUCUUUUCCAGCUUCCAGGCGCAGUAGGCC .((((((((...(((((((.....(((((..(((.(((((.(..((((....))))..).)))..........)))))..)))))..)))))))......(((....))))))).)))). ( -32.10) >DroYak_CAF1 1846 120 + 1 AGUUUCUGCUUCAGCUUCUCCAGCUGCUGCUGAAAGCUCUCGUUGAGCUGCAGCUGCUCUAGACAAUCCAAGUGUUCCUCCAGGGAAGCAAUAUUUUUCAGCUCCCAGGCGCAGUAGGCC .((((((((....(((((.(((((.(((((....(((((.....)))))))))).)))...((((.......))))......)))))))...........(((....))))))).)))). ( -38.70) >DroAna_CAF1 1710 120 + 1 AAUUCGUUCUUUAAGUUUUCCAAUUGCUGAAGAAAAGAUUCAUUUAAUUGCAUCUGCUCCAAGCUGGAGAGAUGCUUUUCCAGGACAUCCAGGUCCUCUAGCUUCCAGGCGCAAUGUGCC ((((..(((((((.((.........)))))))))..)))).........((((((.((((.....))))))))))......(((((......)))))..........(((((...))))) ( -31.50) >consensus AGUUUCUGCUUCAGGUCUUCCAGCUGCUGCUGAAAGCUCUCCUUAAGCUGCAGCUGCCCGAGACAAUCCAAAUGCUCCUCCAGCGAAGCAAUCUUCUUCAGCUUCCAGGCGCAGUAGGCC ....(((((.....((((..(((((((.(((.((........)).))).)))))))....))))........(((.(((.....(((((...........))))).))).)))))))).. (-19.77 = -20.70 + 0.93)


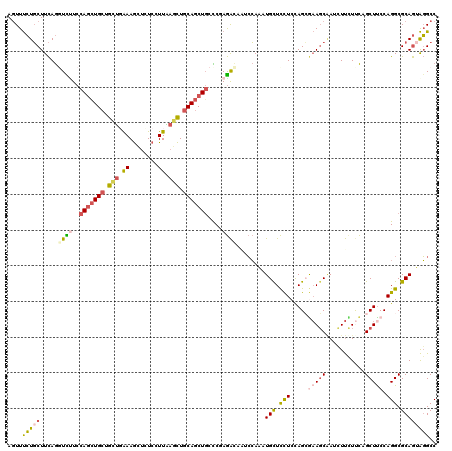
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:02 2006