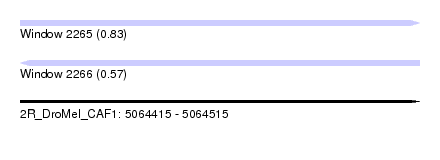
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,064,415 – 5,064,515 |
| Length | 100 |
| Max. P | 0.833970 |

| Location | 5,064,415 – 5,064,515 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -21.37 |
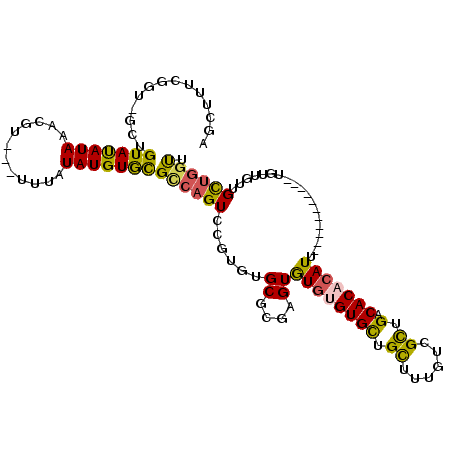
| Consensus MFE | -13.10 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.833970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5064415 100 + 20766785 AGCCAGCAACAACA-----------AAUGUGUGUCAGCGACAAAGCAGCACACACACUCGCGCACACUGACUGGCGCACAUAUAAA---ACGUUUAUAUACAGC-ACCAAAAGCU .(((((((......-----------..((((((((.((......)).).)))))))...........)).)))))((..((((((.---....))))))...))-.......... ( -21.81) >DroSec_CAF1 7253 100 + 1 AACCAGCAACAACA-----------AAUGUGUGUCAGCGACAAAGCAGCACACACACUCGCGCACACGGACUGGCGCACAUAUAAA---ACGUUUAUAUACAGC-AUCGAAAGCU ....(((.......-----------..((((((((.((......)).).))))))).((((((.((.....))))))..((((((.---....)))))).....-...))..))) ( -22.00) >DroSim_CAF1 7381 100 + 1 AACCAGCAACAACA-----------AAUGUGUGUCAGCGACAAAGCUGCACACACACUCGCGCACACGGACUGGCGCACAUAUAAA---ACGUUUAUAUACAGC-ACCGAAAGCU ....(((.......-----------..(((((((((((......)))).))))))).((((((.((.....))))))..((((((.---....)))))).....-...))..))) ( -27.30) >DroEre_CAF1 7451 100 + 1 AACCAGCAACAACA-----------AAUGUGUGUCAGCGACAAAGCAGCACUCACACUCGCGCACACGGACUGACGCACAUAUACA---ACGUUUAUAUACAGC-ACAGAAAGCU .....((.......-----------.(((((((((((.......((.((..........)))).......))))))))))).....---..((......)).))-.......... ( -20.44) >DroMoj_CAF1 9314 114 + 1 GAGCAACAGCAACAACCACAAAUUGAAUGUGUGUCAACGACAAAACUACACCAAUACACACGCACACAUACA-GCAUACAUACACAUUCUCUCUUAUGUACGAUUGUAAAGAGCU .(((....((..(((.......)))...((((((.....................)))))))).....((((-((.((((((............)))))).).)))))....))) ( -15.30) >consensus AACCAGCAACAACA___________AAUGUGUGUCAGCGACAAAGCAGCACACACACUCGCGCACACGGACUGGCGCACAUAUAAA___ACGUUUAUAUACAGC_ACCGAAAGCU ..........................(((((((((((.......((.((..........)))).......))))))))))).................................. (-13.10 = -13.82 + 0.72)

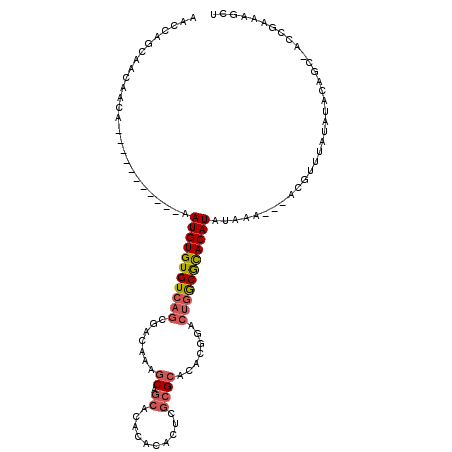
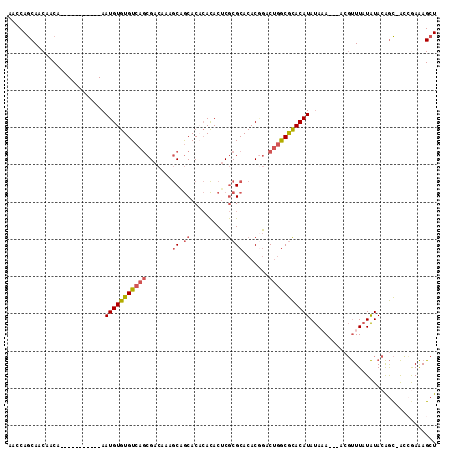
| Location | 5,064,415 – 5,064,515 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -16.20 |
| Energy contribution | -15.84 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5064415 100 - 20766785 AGCUUUUGGU-GCUGUAUAUAAACGU---UUUAUAUGUGCGCCAGUCAGUGUGCGCGAGUGUGUGUGCUGCUUUGUCGCUGACACACAUU-----------UGUUGUUGCUGGCU ......((((-((...((((((....---.))))))..))))))(((((((...((((((((((((.(.((......)).))))))))))-----------)))...))))))). ( -38.10) >DroSec_CAF1 7253 100 - 1 AGCUUUCGAU-GCUGUAUAUAAACGU---UUUAUAUGUGCGCCAGUCCGUGUGCGCGAGUGUGUGUGCUGCUUUGUCGCUGACACACAUU-----------UGUUGUUGCUGGUU (((.......-)))(((((((.....---....)))))))((((((......((((((((((((((.(.((......)).))))))))))-----------)).))).)))))). ( -30.50) >DroSim_CAF1 7381 100 - 1 AGCUUUCGGU-GCUGUAUAUAAACGU---UUUAUAUGUGCGCCAGUCCGUGUGCGCGAGUGUGUGUGCAGCUUUGUCGCUGACACACAUU-----------UGUUGUUGCUGGUU .......(((-((...((((((....---.))))))..)))))...(((...((((((((((((((.((((......)))))))))))))-----------)).)))...))).. ( -35.80) >DroEre_CAF1 7451 100 - 1 AGCUUUCUGU-GCUGUAUAUAAACGU---UGUAUAUGUGCGUCAGUCCGUGUGCGCGAGUGUGAGUGCUGCUUUGUCGCUGACACACAUU-----------UGUUGUUGCUGGUU (((.......-)))(((.((((((((---.(((((((..(....)..)))))))))).((((.(((((......).)))).))))...))-----------)))...)))..... ( -26.00) >DroMoj_CAF1 9314 114 - 1 AGCUCUUUACAAUCGUACAUAAGAGAGAAUGUGUAUGUAUGC-UGUAUGUGUGCGUGUGUAUUGGUGUAGUUUUGUCGUUGACACACAUUCAAUUUGUGGUUGUUGCUGUUGCUC (((.....(((((((((((((........)))))))((((((-.....))))))((((((..((..(......)..))...))))))...........)))))).)))....... ( -27.20) >consensus AGCUUUCGGU_GCUGUAUAUAAACGU___UUUAUAUGUGCGCCAGUCCGUGUGCGCGAGUGUGUGUGCUGCUUUGUCGCUGACACACAUU___________UGUUGUUGCUGGUU ..............(((((((............)))))))((((((......((....))((((((((.((......)).).)))))))...................)))))). (-16.20 = -15.84 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:44 2006