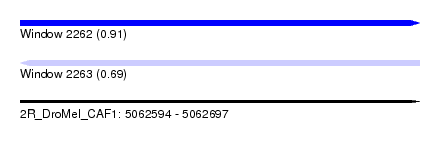
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,062,594 – 5,062,697 |
| Length | 103 |
| Max. P | 0.910223 |

| Location | 5,062,594 – 5,062,697 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -21.52 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
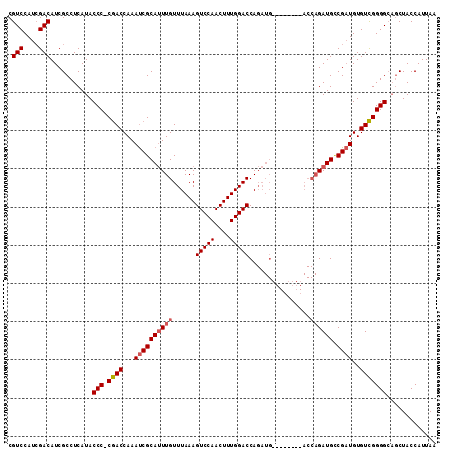
>2R_DroMel_CAF1 5062594 103 + 20766785 CGUCCAUCGACAUCGCCUCAUACCC-CGACCAAAUCGCAUUUGUUUAAAGUCCAACUUUGGACCAGAUG--------ACCAGAUGGCGAUGUGUCGGGGCAGCUACCAUUAA .(((....)))...........(((-((((...((((((((((.(((..(((((....)))))....))--------).))))).)))))..)))))))............. ( -32.70) >DroSec_CAF1 5460 108 + 1 CGUCCAUCGACAUCGCCUCAUACCC-CGACCAAAUCGCAUUUGUUUAAAGUCCAACUUUGGACCAGAUGACCAGAUGACCAGAUGUCGAUGUGUCGGGG---CUACCAUUAA ((.(((((((((((...((((....-(((.....)))(((((((((((((.....))))))).)))))).....))))...)))))))))).).))((.---...))..... ( -33.80) >DroSim_CAF1 5556 103 + 1 CGUCCAUCGACAUCGCCUCAUACCC-CGACCAAACCGCAUUUGUUUAAAGUCCAACUUUGGACCAGAUG--------ACCAGAUGCCGCUGUGUCGGGGCAGCUACCAUUAA .(((....)))...........(((-((((((....(((((((.(((..(((((....)))))....))--------).)))))))...)).)))))))............. ( -30.70) >DroEre_CAF1 5779 96 + 1 CGUCCAUAGACAUCGCCUCAUACCCCCAACCAAAUCGCAAUUGUUUAAAGUCCAACUUUGGACCA----------------GAUGCCGAUGUGUUGGGGCAGCUACCAUUAG .(((....)))............(((((((...((((((.((((((((((.....))))))).))----------------).)).))))..)))))))............. ( -25.90) >consensus CGUCCAUCGACAUCGCCUCAUACCC_CGACCAAAUCGCAUUUGUUUAAAGUCCAACUUUGGACCAGAUG________ACCAGAUGCCGAUGUGUCGGGGCAGCUACCAUUAA .(((....)))...........(((.((((...((((((((((......(((((....)))))................)))))).))))..)))))))............. (-21.52 = -22.15 + 0.63)

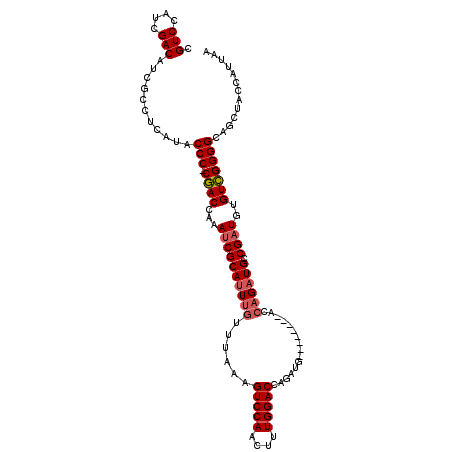
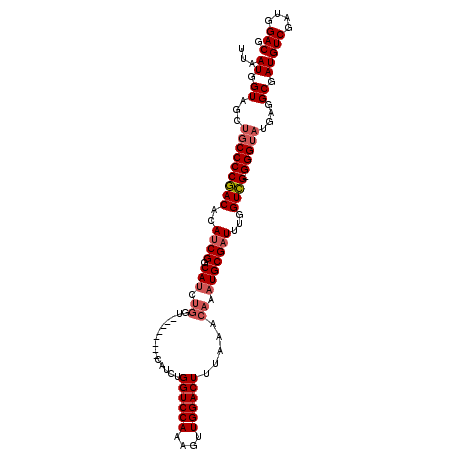
| Location | 5,062,594 – 5,062,697 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.35 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5062594 103 - 20766785 UUAAUGGUAGCUGCCCCGACACAUCGCCAUCUGGU--------CAUCUGGUCCAAAGUUGGACUUUAAACAAAUGCGAUUUGGUCG-GGGUAUGAGGCGAUGUCGAUGGACG ...((.((..((((((((((..(((((.((.((..--------.....((((((....)))))).....)).)))))))...))))-))))).)..)).))(((....))). ( -37.80) >DroSec_CAF1 5460 108 - 1 UUAAUGGUAG---CCCCGACACAUCGACAUCUGGUCAUCUGGUCAUCUGGUCCAAAGUUGGACUUUAAACAAAUGCGAUUUGGUCG-GGGUAUGAGGCGAUGUCGAUGGACG ...((.((.(---(((((((..((((.(((.((..(....)..))...((((((....))))))........)))))))...))))-)))).....)).))(((....))). ( -33.80) >DroSim_CAF1 5556 103 - 1 UUAAUGGUAGCUGCCCCGACACAGCGGCAUCUGGU--------CAUCUGGUCCAAAGUUGGACUUUAAACAAAUGCGGUUUGGUCG-GGGUAUGAGGCGAUGUCGAUGGACG ...((.((..((((((((((..(((.((((.((..--------.....((((((....)))))).....)).)))).)))..))))-))))).)..)).))(((....))). ( -40.10) >DroEre_CAF1 5779 96 - 1 CUAAUGGUAGCUGCCCCAACACAUCGGCAUC----------------UGGUCCAAAGUUGGACUUUAAACAAUUGCGAUUUGGUUGGGGGUAUGAGGCGAUGUCUAUGGACG ...((.((..(((((((..((.((((.((..----------------.((((((....)))))).........)))))).))....)))))).)..)).))(((....))). ( -27.90) >consensus UUAAUGGUAGCUGCCCCGACACAUCGGCAUCUGGU________CAUCUGGUCCAAAGUUGGACUUUAAACAAAUGCGAUUUGGUCG_GGGUAUGAGGCGAUGUCGAUGGACG ...((.((...(((((((((..((((.(((.((...............((((((....)))))).....)).)))))))...)))).)))))....)).))(((....))). (-25.23 = -26.35 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:41 2006