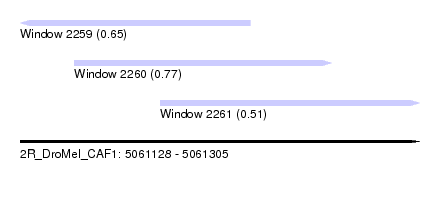
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,061,128 – 5,061,305 |
| Length | 177 |
| Max. P | 0.773306 |

| Location | 5,061,128 – 5,061,230 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5061128 102 - 20766785 UUUUCCCCGAAUGCGAUUGCAUUGCCAGGGGAGGUUAUUCUGAACUGAAACCAGUUUGAUGGCUUUUCCCUAUAUGU--GCA----------------AGUACAUUCCAUAGUUUCCUGU ........(((((...((((((....(((((((((((((..((((((....)))))))))))).)))))))....))--)))----------------)...)))))............. ( -30.40) >DroSec_CAF1 4023 99 - 1 UUUUCCCCGAAUACGAUGGCAAUGCCAGGGGAGGUUUUUCUGAACUGGAACCAGUUUGGUGGCUUUUCCCUAAAU----GCA----------------CGUACAUU-CAUAGGUUCCUGU ........(((((((.((((...))))((..(((((...(..(((((....)))))..).)))))..))......----...----------------))))..))-)((((....)))) ( -29.30) >DroSim_CAF1 4091 99 - 1 UUUUCCCCGAAUACGAUUGCAUUGCCAGGGGAGGUUAUUCUGAACUGGAACCAGUUUGGUGGCUUUUCCCUAUAU----GCA----------------CGUACAUU-CAUAGGUUCCUGU ........(((((((..(((((....(((((((((((..(..(((((....)))))..))))).)))))))..))----)))----------------))))..))-)((((....)))) ( -29.90) >DroEre_CAF1 4208 119 - 1 UUUUCCCCGCACACAAUUGCAAUGCCCGGGGAGGUUAUUUUGAACUGAAACCAGUUUGGUGGCUUUUCCCUAUUUGUAUGCACAUGCAUACAUACAUAUGUACAUU-CAAAGGCUCCUGU ........(((......)))...(((.((..((((((((..((((((....))))))))))))))..)).....(((((((....)))))))..............-....)))...... ( -32.50) >consensus UUUUCCCCGAAUACGAUUGCAAUGCCAGGGGAGGUUAUUCUGAACUGAAACCAGUUUGGUGGCUUUUCCCUAUAU____GCA________________CGUACAUU_CAUAGGUUCCUGU ..................(((..(((.((..((((((((..((((((....))))))))))))))..))..........((..................))..........)))...))) (-17.59 = -17.72 + 0.13)

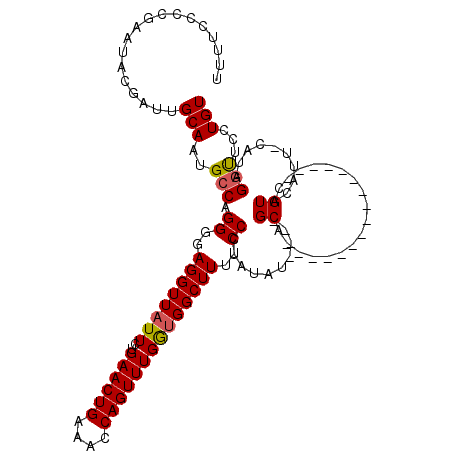
| Location | 5,061,152 – 5,061,266 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5061152 114 + 20766785 C--ACAUAUAGGGAAAAGCCAUCAAACUGGUUUCAGUUCAGAAUAACCUCCCCUGGCAAUGCAAUCGCAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCGGAAGCCCCUUGAU .--((((....(((.((((((......))))))...))).........(((((....(((((....))))).)))))........))-)).....((((..((....)).))))... ( -27.20) >DroSec_CAF1 4046 112 + 1 C----AUUUAGGGAAAAGCCACCAAACUGGUUCCAGUUCAGAAAAACCUCCCCUGGCAUUGCCAUCGUAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCAGAAGCCCCUUGAU .----.....(((....((..((.(((((....)))))..........(((((((((...))))..(....))))))..........-.........))..)).....)))...... ( -23.80) >DroSim_CAF1 4114 112 + 1 C----AUAUAGGGAAAAGCCACCAAACUGGUUCCAGUUCAGAAUAACCUCCCCUGGCAAUGCAAUCGUAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCGGAAGCCCCUUGAU .----....((((........((.(((((....)))))..........(((((....(((((....))))).)))))..........-.........))..((....)))))).... ( -26.10) >DroEre_CAF1 4247 117 + 1 CAUACAAAUAGGGAAAAGCCACCAAACUGGUUUCAGUUCAAAAUAACCUCCCCGGGCAUUGCAAUUGUGUGCGGGGAAAAUAAAAAUAGUAAACAAAGGAAGCGGAAGCCCCUUGAU ..(((......(((.((((((......))))))...))).........((((((.((((.......)))).))))))...........)))....((((..((....)).))))... ( -31.60) >consensus C____AUAUAGGGAAAAGCCACCAAACUGGUUCCAGUUCAGAAUAACCUCCCCUGGCAAUGCAAUCGUAUUCGGGGAAAAUAAAAAU_GUAAACAAAGGAAGCGGAAGCCCCUUGAU .........((((.....((....(((((....)))))..........(((((....(((((....))))).)))))....................))..((....)))))).... (-22.70 = -23.08 + 0.37)

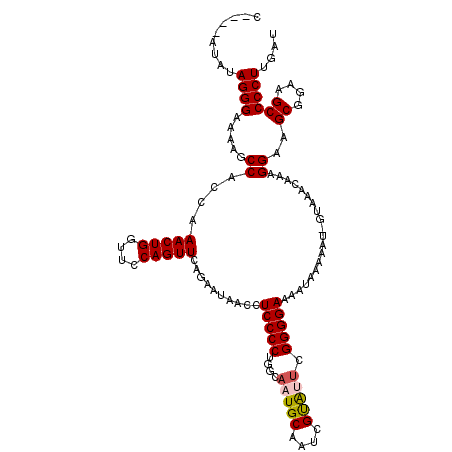
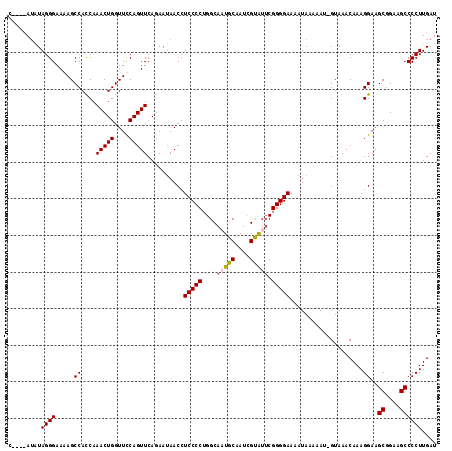
| Location | 5,061,190 – 5,061,305 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5061190 115 + 20766785 GAAUAACCUCCCCUGGCAAUGCAAUCGCAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCGGAAGCCCCUUGAUAAGGCGCAAUUUCUUGCCAAGUUAUCAUUCUAAUGAAAA ((((....(((((....(((((....))))).)))))..........-.......((((..((....)).))))(((((((((........))))...)))))))))......... ( -27.10) >DroSec_CAF1 4082 115 + 1 GAAAAACCUCCCCUGGCAUUGCCAUCGUAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCAGAAGCCCCUUGAUAAGGCGCCAUUUCUUGCCAAGUUAUCAUUCUAAUGAAAA ........(((((((((...))))..(....)))))).......(((-(..(((.((((..((....)).)))).....((((........))))..)))..)))).......... ( -21.50) >DroSim_CAF1 4150 115 + 1 GAAUAACCUCCCCUGGCAAUGCAAUCGUAUUCGGGGAAAAUAAAAAU-GUAAACAAAGGAAGCGGAAGCCCCUUGAUAAGGCGCAAUUUCUUGCCAAGUUAUCAUUCUAAUGAAAA ((((....(((((....(((((....))))).)))))..........-.......((((..((....)).))))(((((((((........))))...)))))))))......... ( -25.10) >DroEre_CAF1 4287 115 + 1 AAAUAACCUCCCCGGGCAUUGCAAUUGUGUGCGGGGAAAAUAAAAAUAGUAAACAAAGGAAGCGGAAGCCCCUUGAUAAGGCGCAAUUUCUUGCCAAGUUAUCAUUCUAAUGAAA- ........((((((.((((.......)))).))))))..................((((..((....)).))))(((((((((........))))...))))).(((....))).- ( -31.20) >consensus GAAUAACCUCCCCUGGCAAUGCAAUCGUAUUCGGGGAAAAUAAAAAU_GUAAACAAAGGAAGCGGAAGCCCCUUGAUAAGGCGCAAUUUCUUGCCAAGUUAUCAUUCUAAUGAAAA ........(((((....(((((....))))).)))))..................((((..((....)).))))(((((((((........))))...))))).(((....))).. (-22.05 = -22.42 + 0.38)

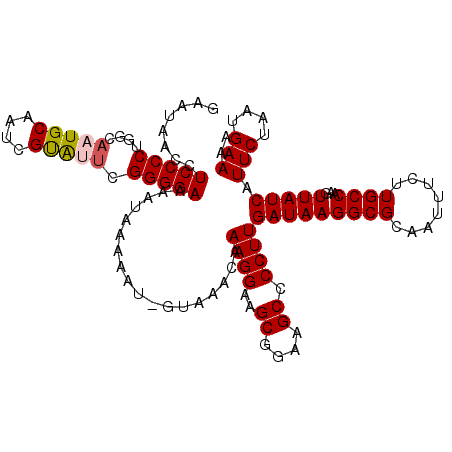
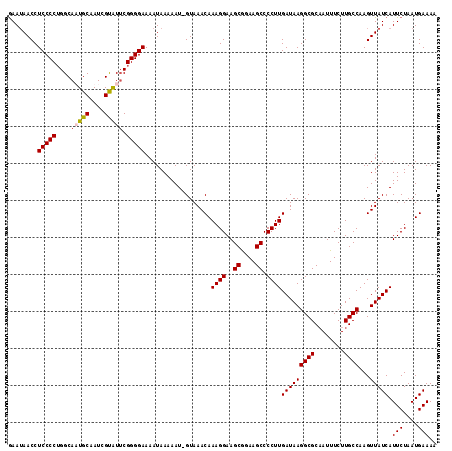
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:40 2006