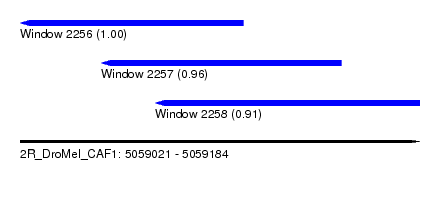
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,059,021 – 5,059,184 |
| Length | 163 |
| Max. P | 0.999758 |

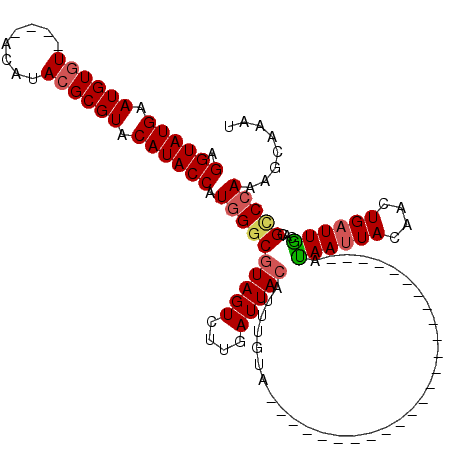
| Location | 5,059,021 – 5,059,112 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5059021 91 - 20766785 AGGUAUGAAUGUCU----ACAUACGCGUACAUACCAUGGACGUAGUCUUGAUUACAUUUGUA-------------------------AUAAUUACAACUGAUUACACGCCCAAACCAAAU .((((((.((((..----......)))).)))))).(((.(((.((...(((((...(((((-------------------------.....))))).)))))))))).)))........ ( -18.90) >DroSec_CAF1 1876 91 - 1 AGGUAUGAAUGUGU----ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA-------------------------AUAAUUACAUCUGAUUGCACGUCCAAAGCAAAU .((((((.((((((----....)))))).)))))).(((((((((((.(((((((....)))-------------------------)).....))...))))..)))))))........ ( -27.10) >DroSim_CAF1 1888 91 - 1 AGGUAUGAAUGUGU----ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA-------------------------AUAAUUACAACUGAUUGCACGUCCAAAGCAAAU .((((((.((((((----....)))))).)))))).(((((((.((...(((((...(((((-------------------------.....))))).))))))))))))))........ ( -27.60) >DroEre_CAF1 1893 120 - 1 AGGUAUGAAUGUGUACGUACAUACGCGUACAUACCAUGGGCGUAGUAGUGAUUAGAUUGGUGGGUAUUAUGUACAUAUUUGUGUGGUACAAGUACAACUGAUUGCGUGCCAAAAGAAAAU .((((((.(((((((......))))))).))))))...((((.....(..(((((.(((....((((((..((......))..)))))).....))))))))..).)))).......... ( -36.30) >consensus AGGUAUGAAUGUGU____ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA_________________________AUAAUUACAACUGAUUGCACGCCCAAAGCAAAU .((((((.((((((........)))))).)))))).((((((((((....))))).................................((((((....))))))...)))))........ (-20.10 = -20.85 + 0.75)


| Location | 5,059,054 – 5,059,152 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -23.73 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5059054 98 - 20766785 AUGAAAACAGUACAACGCUGCAUUACACACAUGCCAGCUCAGGUAUGAAUGUCU----ACAUACGCGUACAUACCAUGGACGUAGUCUUGAUUACAUUUGUA------------------ ......(((((.((..(((((((.......))).))))...((((((.((((..----......)))).)))))).)).))(((((....)))))...))).------------------ ( -21.60) >DroSec_CAF1 1909 98 - 1 AUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGU----ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA------------------ .......((((.....))))...((((.........(((((((((((.((((((----....)))))).)))))).)))))(((((....)))))...))))------------------ ( -28.30) >DroSim_CAF1 1921 98 - 1 AUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGU----ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA------------------ .......((((.....))))...((((.........(((((((((((.((((((----....)))))).)))))).)))))(((((....)))))...))))------------------ ( -28.30) >DroEre_CAF1 1933 120 - 1 ACGAAAGCACUAAAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGUACGUACAUACGCGUACAUACCAUGGGCGUAGUAGUGAUUAGAUUGGUGGGUAUUAUGUACAUAUUU .....(((........)))((((...((...((((((((((((((((.(((((((......))))))).)))))).))))).((((....))))...))))).))...))))........ ( -34.10) >consensus AUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGU____ACAUACGCGUACAUACCAUGGGCGUAGUCUUGAUUACAUUUGUA__________________ .........((((((.(((((....(......)...(((((((((((.((((((........)))))).)))))).)))))))))).)))..)))......................... (-23.73 = -24.35 + 0.62)


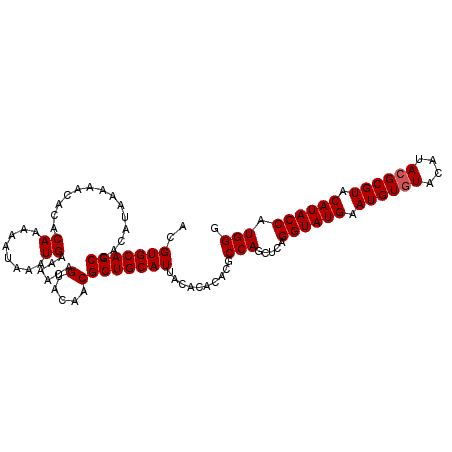
| Location | 5,059,076 – 5,059,184 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5059076 108 - 20766785 ACGUGCAGCCACAGAAGAACACACAAAAAUAAAUGAAAACAGUACAACGCUGCAUUACACACAUGCCAGCUCAGGUAUGAAUGUCUACAUACGCGUACAUACCAUGGA ..(((((((..............((........))......(.....))))))))..........(((.....((((((.((((........)))).)))))).))). ( -19.30) >DroSec_CAF1 1931 108 - 1 ACGUGCAGCCACAUAAAAACACACAAAAAUAAAUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGUACAUACGCGUACAUACCAUGGG ..(((((((..............((........))......(.....))))))))..............((((((((((.((((((....)))))).)))))).)))) ( -24.40) >DroSim_CAF1 1943 108 - 1 ACGUGCAGCCACAUGAAAACACACAAAAAUAAAUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGUACAUACGCGUACAUACCAUGGG ..(((((((..(((..................)))......(.....))))))))..............((((((((((.((((((....)))))).)))))).)))) ( -25.07) >consensus ACGUGCAGCCACAUAAAAACACACAAAAAUAAAUGAAAACAGUACAACGCUGCAUUACACACACGCCAGCUCAGGUAUGAAUGUGUACAUACGCGUACAUACCAUGGG ..(((((((..............((........))......(.....))))))))..........(((.....((((((.((((((....)))))).)))))).))). (-21.90 = -22.23 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:37 2006