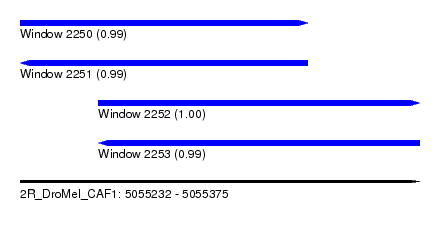
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,055,232 – 5,055,375 |
| Length | 143 |
| Max. P | 0.995919 |

| Location | 5,055,232 – 5,055,335 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -16.73 |
| Energy contribution | -18.35 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5055232 103 + 20766785 UUAAGCUAUCUUAGAGACUGAAAAACUA-----------------UUUCACAAUCUCUGUUUAACAGAGAACGAACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUC .((((((...(((((....((((.....-----------------........((((((.....)))))).....((((......))))......))))...)))))..))))))..... ( -22.70) >DroSec_CAF1 9767 91 + 1 UUAAGCUAUC--------UGGAAAACUA-----------------UUUCCCAAUCUCUGUUUAACAGAGA----ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUC .((((((..(--------((((......-----------------........((((((.....))))))----.((((......)))).............)))))..))))))..... ( -22.00) >DroSim_CAF1 6795 99 + 1 UUAAGCUAUCUUAGAGACUGGAAAACUA-----------------UUUCCCAAUCUCUGUUUAACAGAGA----ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGCGUUUAUCUUC ....((((....(((((.(((.......-----------------........((((((.....))))))----.((((......))))..))).)))))....))))............ ( -24.50) >DroEre_CAF1 13865 116 + 1 UGAUGAUAUCUUAGAGACUGGAAAACCAUAUUUUAGGCCAAUAAAUUUCCCAAUCUGUGUUUAACACAGA----ACCACAUAGAAGUGGCCCCAAUUUAUCAACUAGCGGGUUUAUCUUU .(((((...(((((((..(((....)))..)))))))...........(((..((((((.....))))))----.((((......))))...................))).)))))... ( -24.60) >consensus UUAAGCUAUCUUAGAGACUGGAAAACUA_________________UUUCCCAAUCUCUGUUUAACAGAGA____ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUC ....((((....(((((.(((................................((((((.....)))))).....((((......))))..))).)))))....))))............ (-16.73 = -18.35 + 1.62)

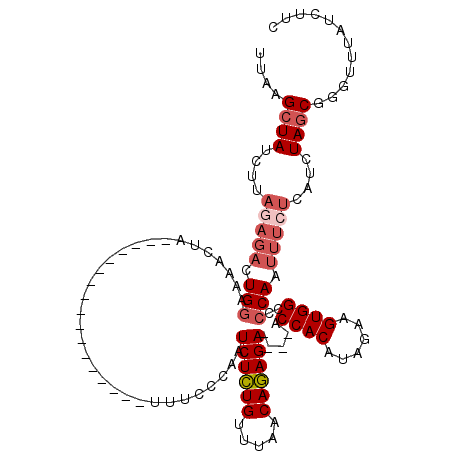
| Location | 5,055,232 – 5,055,335 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -22.31 |
| Energy contribution | -23.50 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5055232 103 - 20766785 GAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGUUCGUUCUCUGUUAAACAGAGAUUGUGAAA-----------------UAGUUUUUCAGUCUCUAAGAUAGCUUAA ............(((...(((((((((((((((((......)))))))..((((((.....))))))........-----------------))))))))))(((.....)))))).... ( -29.10) >DroSec_CAF1 9767 91 - 1 GAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU----UCUCUGUUAAACAGAGAUUGGGAAA-----------------UAGUUUUCCA--------GAUAGCUUAA ............((((..((..((((((..(((((......)))))----((((((.....))))))........-----------------))))))..))--------..)))).... ( -26.30) >DroSim_CAF1 6795 99 - 1 GAAGAUAAACGCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU----UCUCUGUUAAACAGAGAUUGGGAAA-----------------UAGUUUUCCAGUCUCUAAGAUAGCUUAA ............((((.......(((.((((((((......)))))----)))..)))....((((((((((((.-----------------....))))))))))))....)))).... ( -30.40) >DroEre_CAF1 13865 116 - 1 AAAGAUAAACCCGCUAGUUGAUAAAUUGGGGCCACUUCUAUGUGGU----UCUGUGUUAAACACAGAUUGGGAAAUUUAUUGGCCUAAAAUAUGGUUUUCCAGUCUCUAAGAUAUCAUCA ................(.(((((....((((((((......)))))----)))(((.....)))(((((((((((((((((.......)))).)))))))))))))......))))).). ( -30.80) >consensus GAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU____UCUCUGUUAAACAGAGAUUGGGAAA_________________UAGUUUUCCAGUCUCUAAGAUAGCUUAA ............((((....(((.(((((((((((......)))))....((((((.....))))))...............................)))))).)))....)))).... (-22.31 = -23.50 + 1.19)

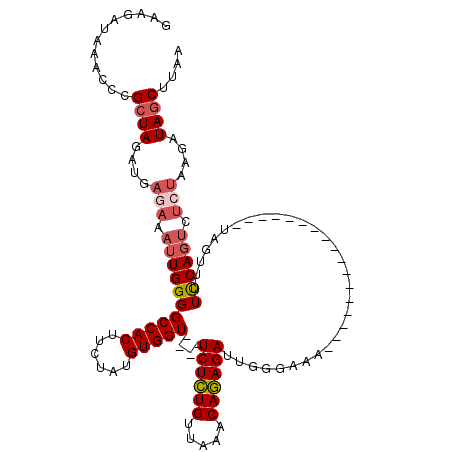
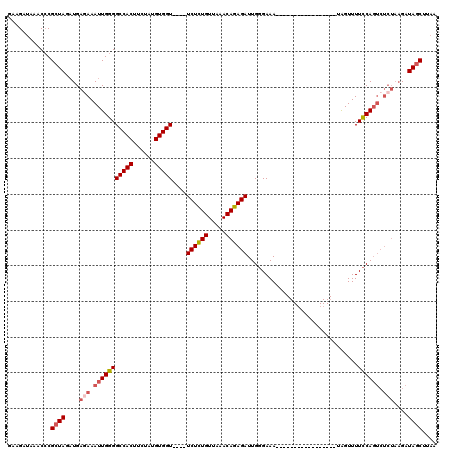
| Location | 5,055,260 – 5,055,375 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -25.99 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
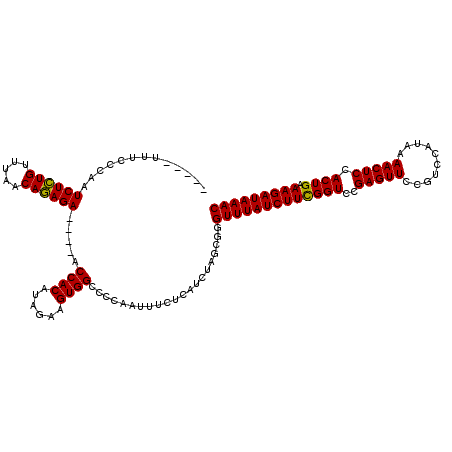
>2R_DroMel_CAF1 5055260 115 + 20766785 -----UUUCACAAUCUCUGUUUAACAGAGAACGAACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUCGGUCCGAGUUCCGUCCAUAAAACUCCACUGAAAGAUAAAC -----........((((((.....)))))).....((.(.((((.(.((........)).).))))).))(((((((((((((..(((((..........))))).)))).))))))))) ( -27.00) >DroSec_CAF1 9787 111 + 1 -----UUUCCCAAUCUCUGUUUAACAGAGA----ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUCGGUCCGAGUUCCGUCCAUAAAACUCCACUGAAAGAUAAAC -----...(((..((((((.....))))))----.((((......))))...................)))((((((((((((..(((((..........))))).)))).)))))))). ( -29.40) >DroSim_CAF1 6823 111 + 1 -----UUUCCCAAUCUCUGUUUAACAGAGA----ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGCGUUUAUCUUCGGUCCGAGUUCCGUCCAUAAAACUCCACUGAAAGAUAAAC -----........((((((.....))))))----.((((......)))).....................(((((((((((((..(((((..........))))).)))).))))))))) ( -27.00) >DroEre_CAF1 13905 116 + 1 AUAAAUUUCCCAAUCUGUGUUUAACACAGA----ACCACAUAGAAGUGGCCCCAAUUUAUCAACUAGCGGGUUUAUCUUUGGUCCGAGUUCCGUCCACAAAACUCCACUGAAAGAUAAAC ........(((..((((((.....))))))----.((((......))))...................)))((((((((((((..(((((..(....)..))))).))).))))))))). ( -28.00) >consensus _____UUUCCCAAUCUCUGUUUAACAGAGA____ACCACAUAGAAGUGGCCCCAAUUUCUCAUCUAGCGGGUUUAUCUUCGGUCCGAGUUCCGUCCAUAAAACUCCACUGAAAGAUAAAC .............((((((.....)))))).....((((......)))).....................(((((((((((((..(((((..........))))).)))).))))))))) (-25.99 = -25.43 + -0.56)

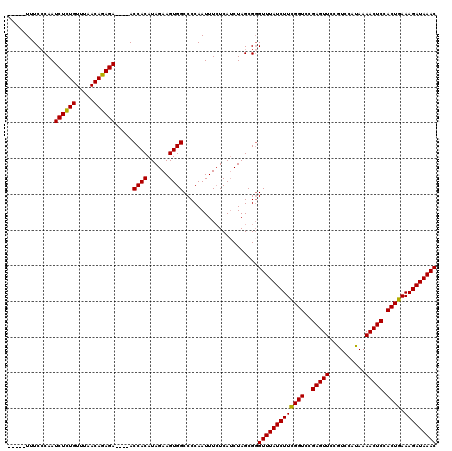
| Location | 5,055,260 – 5,055,375 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -31.94 |
| Energy contribution | -31.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
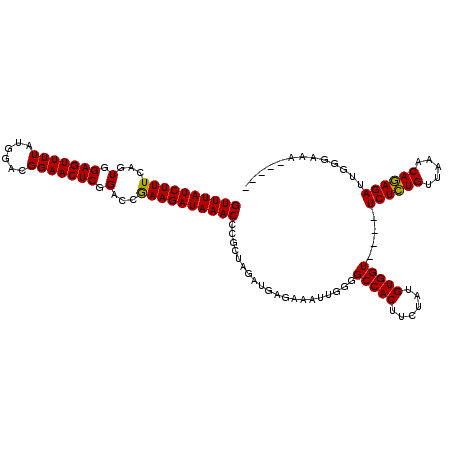
>2R_DroMel_CAF1 5055260 115 - 20766785 GUUUAUCUUUCAGUGGAGUUUUAUGGACGGAACUCGGACCGAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGUUCGUUCUCUGUUAAACAGAGAUUGUGAAA----- ((((((((((...(.(((((((......))))))).)...)))))))))).(((..............(((((((......)))))))..((((((.....))))))..)))...----- ( -36.90) >DroSec_CAF1 9787 111 - 1 GUUUAUCUUUCAGUGGAGUUUUAUGGACGGAACUCGGACCGAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU----UCUCUGUUAAACAGAGAUUGGGAAA----- .(((((((((...(.(((((((......))))))).)...)))))))))(((..................(((((......)))))----((((((.....))))))..)))...----- ( -33.80) >DroSim_CAF1 6823 111 - 1 GUUUAUCUUUCAGUGGAGUUUUAUGGACGGAACUCGGACCGAAGAUAAACGCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU----UCUCUGUUAAACAGAGAUUGGGAAA----- ((((((((((...(.(((((((......))))))).)...))))))))))...((((.............(((((......)))))----((((((.....))))))))))....----- ( -33.40) >DroEre_CAF1 13905 116 - 1 GUUUAUCUUUCAGUGGAGUUUUGUGGACGGAACUCGGACCAAAGAUAAACCCGCUAGUUGAUAAAUUGGGGCCACUUCUAUGUGGU----UCUGUGUUAAACACAGAUUGGGAAAUUUAU .(((((((((...(.((((((((....)))))))).)...)))))))))(((.((((((....)))))).(((((......)))))----((((((.....))))))..)))........ ( -38.90) >consensus GUUUAUCUUUCAGUGGAGUUUUAUGGACGGAACUCGGACCGAAGAUAAACCCGCUAGAUGAGAAAUUGGGGCCACUUCUAUGUGGU____UCUCUGUUAAACAGAGAUUGGGAAA_____ ((((((((((...(.(((((((......))))))).)...))))))))))....................(((((......)))))....((((((.....))))))............. (-31.94 = -31.38 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:32 2006