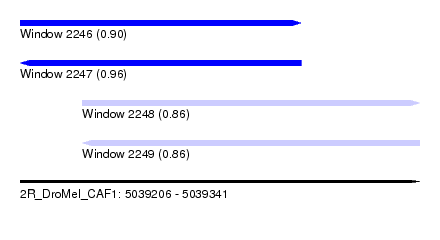
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,039,206 – 5,039,341 |
| Length | 135 |
| Max. P | 0.960018 |

| Location | 5,039,206 – 5,039,301 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -21.95 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
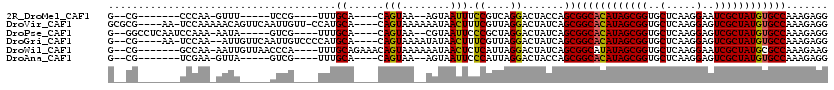
>2R_DroMel_CAF1 5039206 95 + 20766785 G--CG-------CCCAA-GUUU-----UCCG----UUUGCA----CAGUAA--AGUAAUUUCCGUCAGGACUACCAGCGGCACAUAGCGGUGCUCAAGGAAUCGCUAUGUGCCAAAGAGG (--(.-------....(-((((-----(.((----.((((.----......--.))))....))..))))))....))(((((((((((((.(.....).)))))))))))))....... ( -29.40) >DroVir_CAF1 17106 110 + 1 GCGCG----AA-UCCAAAAACAGUUCAAUUGUU-CCAUGCA----CAGUAAAAAAUAACUUUCGUUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGG ((..(----(.-(((............(((((.-......)----))))......((((....)))))))...)).))(((((((((((..((((...)))))))))))))))....... ( -31.00) >DroPse_CAF1 13409 102 + 1 G--GGCCUCAAUCCAAA-AAUA-----GUCG----UUUGCA----CAGUAA--CGUAAUUCCCGCUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGG .--..((((........-.(((-----(((.----(..((.----......--..........)).).))))))....(((((((((((..((((...)))))))))))))))...)))) ( -34.53) >DroGri_CAF1 17451 107 + 1 G--CG----AA-UCCAA--AUUGUUCAAUUGUCCCCAUGCA----CAGUAAAAUAUAACUUUCGUUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGG (--((----((-(....--...))))....((((..(((..----.(((........)))..)))..)))).....))(((((((((((..((((...)))))))))))))))....... ( -32.80) >DroWil_CAF1 14312 106 + 1 G--CG-------GCCAA-AAUUGUUAACCCA----UUUGCAGAAACAGUAAAAAAUAACUCUCAUUAGGACUAUCAGCGGCAUAUAGCGGUGCUCAAGGAAUCGCUAUGCGCCAAAGAAG (--((-------(((..-....((((.....----(((((.......)))))...))))........)).))....))(((.(((((((((.(.....).))))))))).)))....... ( -20.34) >DroAna_CAF1 14154 95 + 1 G--CG-------UCGAA-GUUA-----GUCG----UUUGCA----CAGUAA--AGUAAUUCCCAUUAGGACUACCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGG (--((-------.(((.-....-----.)))----..))).----..((..--.(((..(((.....))).)))..))(((((((((((..((((...)))))))))))))))....... ( -29.90) >consensus G__CG_______UCCAA_AAUA_____AUCG____UUUGCA____CAGUAA__AAUAACUCCCGUUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGG ......................................((......(((........))).((....)).......))((((((((((((..(.....)..))))))))))))....... (-21.95 = -21.48 + -0.47)
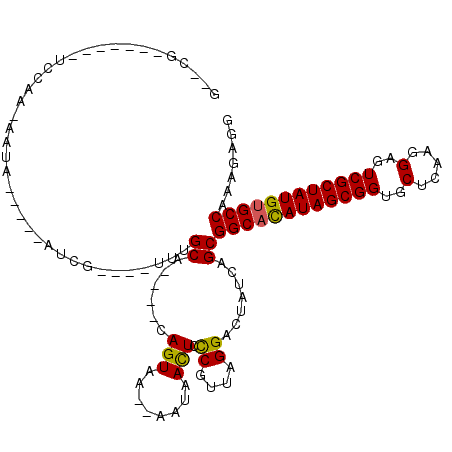
| Location | 5,039,206 – 5,039,301 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.92 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.15 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5039206 95 - 20766785 CCUCUUUGGCACAUAGCGAUUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGAAAUUACU--UUACUG----UGCAAA----CGGA-----AAAC-UUGGG-------CG--C .......(((((((((((.............)))))))))))(((((((.(((....)))...))))--...(((----(....)----))).-----....-...))-------).--. ( -30.32) >DroVir_CAF1 17106 110 - 1 CCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAGUUAUUUUUUACUG----UGCAUGG-AACAAUUGAACUGUUUUUGGA-UU----CGCGC .......(((((((((((.(((...)))...)))))))))))((.((..((((((((....))))..........----.....((-((((.......)))))).)))-))----))).. ( -33.10) >DroPse_CAF1 13409 102 - 1 CCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAGCGGGAAUUACG--UUACUG----UGCAAA----CGAC-----UAUU-UUUGGAUUGAGGCC--C ((((...(((((((((((.(((...)))...)))))))))))...(((((((.(((((.......))--)))..(----(....)----))))-----))))-........))))..--. ( -35.80) >DroGri_CAF1 17451 107 - 1 CCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAGUUAUAUUUUACUG----UGCAUGGGGACAAUUGAACAAU--UUGGA-UU----CG--C ..((..((((((((((((.(((...)))...))))))))))))..)).(((((((((....))))........((----(.((((....))..)).)))..--..)))-))----..--. ( -34.70) >DroWil_CAF1 14312 106 - 1 CUUCUUUGGCGCAUAGCGAUUCCUUGAGCACCGCUAUAUGCCGCUGAUAGUCCUAAUGAGAGUUAUUUUUUACUGUUUCUGCAAA----UGGGUUAACAAUU-UUGGC-------CG--C .......((((.((((((.............)))))).))))((.((((((..((((....))))......))))))...))...----..((((((.....-)))))-------).--. ( -24.62) >DroAna_CAF1 14154 95 - 1 CCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUAAUGGGAAUUACU--UUACUG----UGCAAA----CGAC-----UAAC-UUCGA-------CG--C .......(((((((((((.(((...)))...)))))))))))((.((((((((.....)).))))))--......----.))...----(((.-----....-.))).-------..--. ( -28.90) >consensus CCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAAUUACU__UUACUG____UGCAAA____CGAC_____AAUU_UUGGA_______CG__C .......(((((((((((.(((...)))...)))))))))))((...((((..((((....))))......)))).....))...................................... (-22.48 = -22.15 + -0.33)

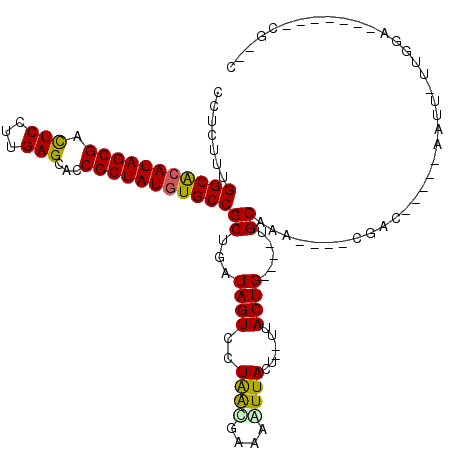
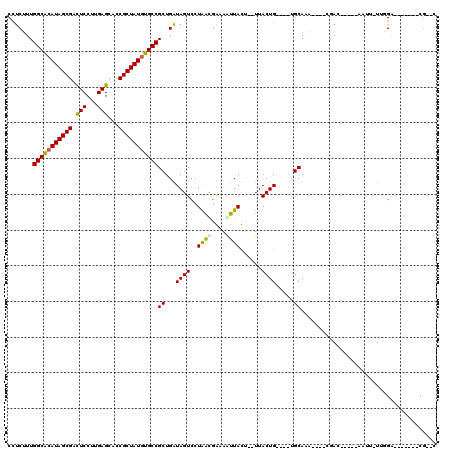
| Location | 5,039,227 – 5,039,341 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -27.75 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5039227 114 + 20766785 A----CAGUAA--AGUAAUUUCCGUCAGGACUACCAGCGGCACAUAGCGGUGCUCAAGGAAUCGCUAUGUGCCAAAGAGGAGCACUACAACAUGCUGCAGACGGACGUCGAGGAGAUGCG .----......--.......((((((.((....)).(((((((((((((((.(.....).))))))))))))).......((((........)))))).))))))((((.....)))).. ( -42.10) >DroPse_CAF1 13437 114 + 1 A----CAGUAA--CGUAAUUCCCGCUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGGAGCACUACAACAUGCUGCAUUCGGACGUGGAGGAGAUGCG .----......--((((.((((..(((.(.((......(((((((((((..((((...))))))))))))))).....(.((((........)))).)....)).).))).)))).)))) ( -39.10) >DroGri_CAF1 17482 116 + 1 A----CAGUAAAAUAUAACUUUCGUUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGGAGCACUACAAUAUGCUGCAGACGGAUGUCGAGGAGAUGCG .----..(((.....((((....)))).(((.(((..((((((((((((..((((...))))))))))))))).....(.((((........)))).).)...)))))).......))). ( -35.90) >DroEre_CAF1 2 93 + 1 ---------------------------GGACUACCAGCGGCACAUAGCGGUGCUCAAGGAGUCGCUAUGUGCCAAAGAGGAGCACUACAACAUGCUGCAGACCGAUGUCGAGGAGAUGCG ---------------------------...((.((.(((((((((((((..((((...))))))))))))))).......((((........)))))).(((....)))..))))..... ( -33.10) >DroWil_CAF1 14338 120 + 1 AGAAACAGUAAAAAAUAACUCUCAUUAGGACUAUCAGCGGCAUAUAGCGGUGCUCAAGGAAUCGCUAUGCGCCAAAGAAGAGCACUACAACAUGCUGCAAACGGAUGUGGAGGAGAUGCG ..................((((((((.((.....).(((((((...(..((((((.......(((...)))........))))))..)...)))))))...).)))).))))........ ( -28.06) >DroMoj_CAF1 2 93 + 1 ---------------------------GGACUAUCAGCGGCACAUAGCGGUACUUAAGGAAUCGCUAUGUGCCAAAGAGGAGCACUACAACAUGCUGCAGACGGACGUGGAGGAGAUGCG ---------------------------...........(((((((((((((.(.....).)))))))))))))........(((((.(..(((((((....))).))))..).)).))). ( -34.00) >consensus A____CAGUAA____UAA_U__CGUUAGGACUAUCAGCGGCACAUAGCGGUGCUCAAGGAAUCGCUAUGUGCCAAAGAGGAGCACUACAACAUGCUGCAGACGGACGUCGAGGAGAUGCG ....................................(((((((((((((((.(.....).))))))))))))).....(.((((........)))).)...................)). (-27.75 = -27.70 + -0.05)
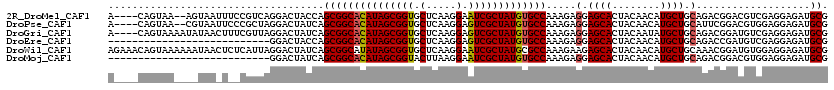
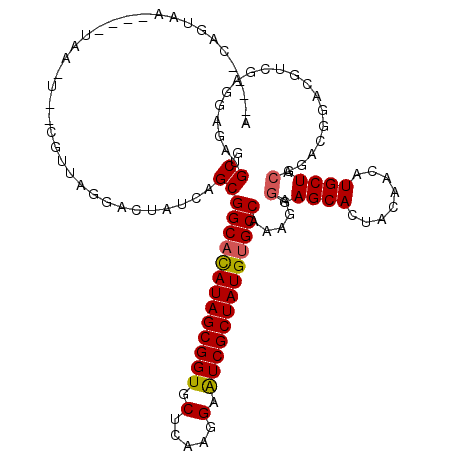
| Location | 5,039,227 – 5,039,341 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
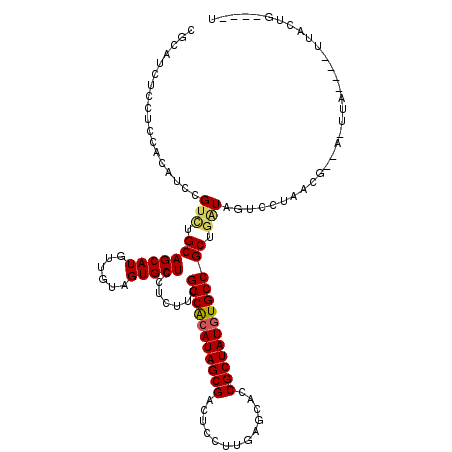
>2R_DroMel_CAF1 5039227 114 - 20766785 CGCAUCUCCUCGACGUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGAUUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCCUGACGGAAAUUACU--UUACUG----U .(((...........((((((.(((((((......))))).......(((((((((((.............))))))))))))).((....)).)))))).......--....))----) ( -36.57) >DroPse_CAF1 13437 114 - 1 CGCAUCUCCUCCACGUCCGAAUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAGCGGGAAUUACG--UUACUG----U .((((.((..........)))))).(((.((((((((..(((.....(((((((((((.(((...)))...)))))))))))((((.......))))))).))))))--..))))----) ( -34.40) >DroGri_CAF1 17482 116 - 1 CGCAUCUCCUCGACAUCCGUCUGCAGCAUAUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACGAAAGUUAUAUUUUACUG----U ...........(((....(((.(((((((......))))).......(((((((((((.(((...)))...))))))))))))).))).))).((((....))))..........----. ( -35.30) >DroEre_CAF1 2 93 - 1 CGCAUCUCCUCGACAUCGGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGGUAGUCC--------------------------- ...........(((((((((((((((....))))))...........(((((((((((.(((...)))...))))))))))))))))).))).--------------------------- ( -34.30) >DroWil_CAF1 14338 120 - 1 CGCAUCUCCUCCACAUCCGUUUGCAGCAUGUUGUAGUGCUCUUCUUUGGCGCAUAGCGAUUCCUUGAGCACCGCUAUAUGCCGCUGAUAGUCCUAAUGAGAGUUAUUUUUUACUGUUUCU .((.((((........(..((.((.((((((.(..((((((.....(.((.....)).)......))))))..).)))))).)).))..).......))))))................. ( -29.16) >DroMoj_CAF1 2 93 - 1 CGCAUCUCCUCCACGUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGAUUCCUUAAGUACCGCUAUGUGCCGCUGAUAGUCC--------------------------- ..............(.(..((.(((((((......))))).......(((((((((((.............))))))))))))).))..).).--------------------------- ( -29.22) >consensus CGCAUCUCCUCCACAUCCGUCUGCAGCAUGUUGUAGUGCUCCUCUUUGGCACAUAGCGACUCCUUGAGCACCGCUAUGUGCCGCUGAUAGUCCUAACG__A_UUA____UUACUG____U ..................(((.(((((((......))))).......(((((((((((.............))))))))))))).)))................................ (-25.62 = -25.70 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:26 2006