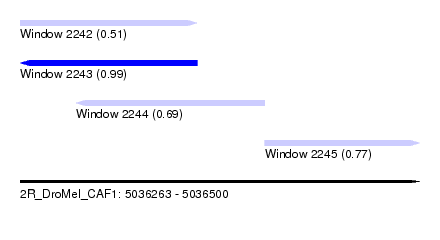
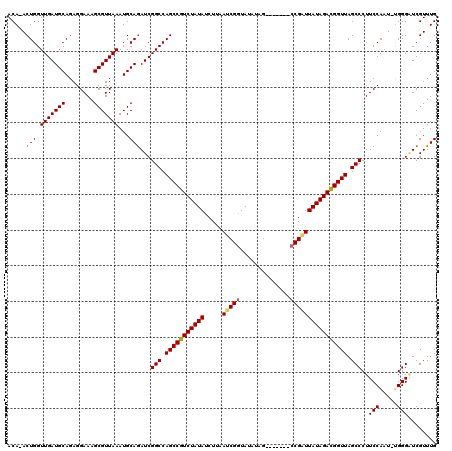
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,036,263 – 5,036,500 |
| Length | 237 |
| Max. P | 0.988795 |

| Location | 5,036,263 – 5,036,368 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5036263 105 + 20766785 -------CAUCCUAUAUAUAUAUAUAUAACAAUCAGAGAGCAAACGAACCCA-AUUGGAAGGGCUAACCAUCUAUAAUCGG-------CUAUAUACCGAUAAAGAUAUAGACGGCUGGCC -------.....((((((....)))))).....................((.-...))...(((((.((.(((((((((((-------.......))))).....)))))).)).))))) ( -20.60) >DroEre_CAF1 16357 101 + 1 -----UGCAUCCUAUA------UAUAAAACAAUCAGAGAGCAAACGAUCCCG-AUUGGAAGGGCUAACCGUCUAUAAGCGG-------CUAUAUACCGAUUAAGAUAUAGACGGCUGGCC -----...........------.......(((((.(.((........))).)-))))....(((((.(((((((((..(((-------.......))).......))))))))).))))) ( -26.50) >DroYak_CAF1 16494 112 + 1 GAAUCAGCAUCCUAUA------UAUA--ACAAUCAGAGAGCAAACGACCCCAAAUUGGAAGGGCUAACCGUCUAUAACCGAUAUUGUACGAUAUAGCGAUGAAGAUAUAGACGGCUGGCC ......((.((((...------....--......)).))))........((.....))...(((((.(((((((((..(..((((((........))))))..).))))))))).))))) ( -25.02) >consensus ______GCAUCCUAUA______UAUA_AACAAUCAGAGAGCAAACGAACCCA_AUUGGAAGGGCUAACCGUCUAUAACCGG_______CUAUAUACCGAUAAAGAUAUAGACGGCUGGCC .................................................((.....))...(((((.(((((((((.............................))))))))).))))) (-16.25 = -16.58 + 0.33)

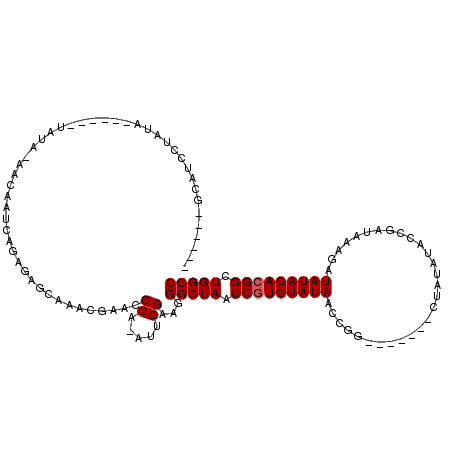
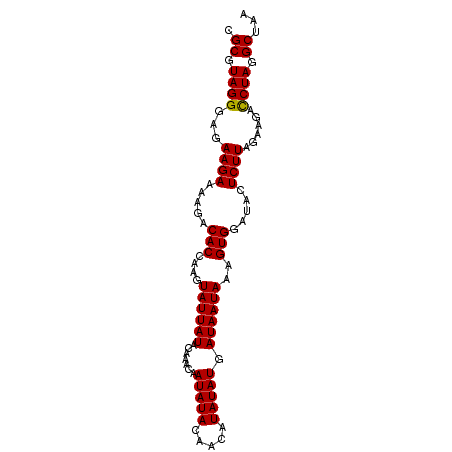
| Location | 5,036,263 – 5,036,368 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5036263 105 - 20766785 GGCCAGCCGUCUAUAUCUUUAUCGGUAUAUAG-------CCGAUUAUAGAUGGUUAGCCCUUCCAAU-UGGGUUCGUUUGCUCUCUGAUUGUUAUAUAUAUAUAUAUAGGAUG------- (((.(((((((((((.....((((((.....)-------)))))))))))))))).)))..(((...-.((((......)))).........((((((....)))))))))..------- ( -32.70) >DroEre_CAF1 16357 101 - 1 GGCCAGCCGUCUAUAUCUUAAUCGGUAUAUAG-------CCGCUUAUAGACGGUUAGCCCUUCCAAU-CGGGAUCGUUUGCUCUCUGAUUGUUUUAUA------UAUAGGAUGCA----- (((.(((((((((((.......((((.....)-------)))..))))))))))).)))....((((-(((((........)).))))))).......------...........----- ( -31.30) >DroYak_CAF1 16494 112 - 1 GGCCAGCCGUCUAUAUCUUCAUCGCUAUAUCGUACAAUAUCGGUUAUAGACGGUUAGCCCUUCCAAUUUGGGGUCGUUUGCUCUCUGAUUGU--UAUA------UAUAGGAUGCUGAUUC ((....))......(((..((((.((((((...(((..(((((...((((((....(((((........)))))))))))....))))))))--...)------)))))))))..))).. ( -28.70) >consensus GGCCAGCCGUCUAUAUCUUAAUCGGUAUAUAG_______CCGAUUAUAGACGGUUAGCCCUUCCAAU_UGGGAUCGUUUGCUCUCUGAUUGUU_UAUA______UAUAGGAUGC______ (((.(((((((((((.....(((((..............)))))))))))))))).)))..(((.....(((........)))...........(((((....))))))))......... (-23.17 = -23.67 + 0.50)

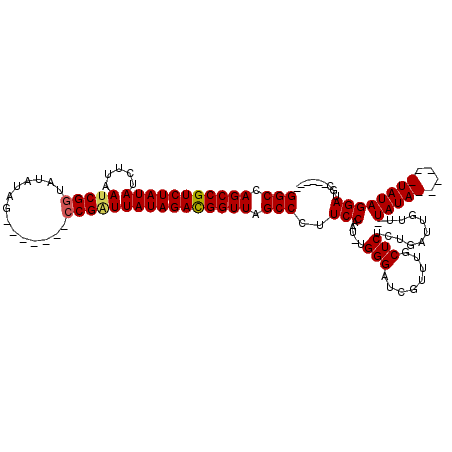
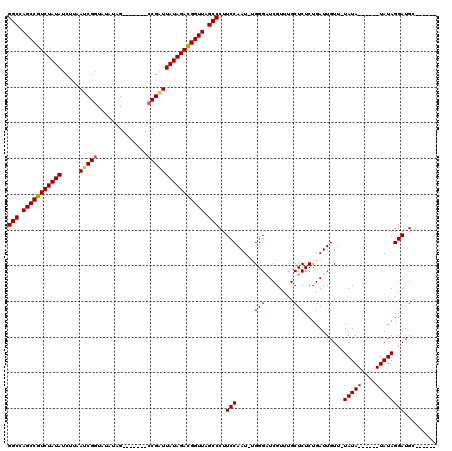
| Location | 5,036,296 – 5,036,408 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.37 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5036296 112 - 20766785 ACAAACUGGUUGAUGCAGAGGAAAGCGUUAAAUGCAAAUCGGCCAGCCGUCUAUAUCUUUAUCGGUAUAUAG-------CCGAUUAUAGAUGGUUAGCCCUUCCAAU-UGGGUUCGUUUG .(((((.((((((((((...((.....))...))))..))))))(((((((((((.....((((((.....)-------))))))))))))))))(((((.......-.))))).))))) ( -38.90) >DroEre_CAF1 16386 111 - 1 ACA-ACUGGUUGAUGCAGAGGAAAGCGUUAAAUGCAGAUCGGCCAGCCGUCUAUAUCUUAAUCGGUAUAUAG-------CCGCUUAUAGACGGUUAGCCCUUCCAAU-CGGGAUCGUUUG ...-.(((((((.((((...((.....))...))))((..(((.(((((((((((.......((((.....)-------)))..))))))))))).)))..))))))-)))......... ( -36.30) >DroYak_CAF1 16526 119 - 1 ACA-ACUGGUUGAUGCAGAGGAAAGCGUUAAAUGCAGAUCGGCCAGCCGUCUAUAUCUUCAUCGCUAUAUCGUACAAUAUCGGUUAUAGACGGUUAGCCCUUCCAAUUUGGGGUCGUUUG ...-.((((((((((((...((.....))...))))..))))))))(((((((((.....((((.(((........))).)))))))))))))...(((((........)))))...... ( -33.50) >consensus ACA_ACUGGUUGAUGCAGAGGAAAGCGUUAAAUGCAGAUCGGCCAGCCGUCUAUAUCUUAAUCGGUAUAUAG_______CCGAUUAUAGACGGUUAGCCCUUCCAAU_UGGGAUCGUUUG .....(((.(((((((........)))))))...)))...(((.(((((((((((.....(((((..............)))))))))))))))).)))..(((.....)))........ (-28.70 = -29.37 + 0.67)

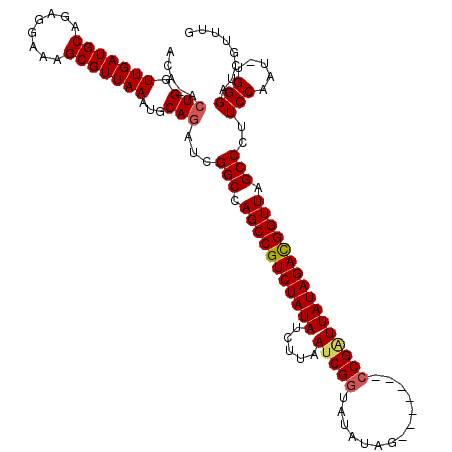
| Location | 5,036,408 – 5,036,500 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -16.57 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5036408 92 + 20766785 CGCGUAGGGAGAAGAUAAGACACCAAGUAUUAUAUAUACAAUAUACAACAUAUAUGAUAAUAAAGUGGAUAUUCUUAGAAGAUCUAGGCUAA .((.((((...((((.....(((....((((((((((............)))))))))).....))).....))))......)))).))... ( -17.70) >DroEre_CAF1 16497 92 + 1 CGCGUAGGGAGAAGAAAUGACACCAAGUAUUAUACAAACAAUAUACAACAUAUAUGAUAAUAAAGUGGAUACUCUUAGAAGACCUAGGCUAA .((.((((...((((.....(((....((((((.......(((((.....))))).))))))..))).....))))......)))).))... ( -15.70) >DroYak_CAF1 16645 92 + 1 CGCGUAGGGAGAAGAAAAGACACCAAGUAUUAUACAAACAAUAUACAACAUAUAUGAUAAUAAAGUGGAUACUCUUAGAAGACCUAGGCUAA .((.((((...((((..(..(((....((((((.......(((((.....))))).))))))..)))..)..))))......)))).))... ( -16.30) >consensus CGCGUAGGGAGAAGAAAAGACACCAAGUAUUAUACAAACAAUAUACAACAUAUAUGAUAAUAAAGUGGAUACUCUUAGAAGACCUAGGCUAA .((.((((...((((.....(((....((((((.......(((((.....))))).))))))..))).....))))......)))).))... (-15.32 = -15.10 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:20 2006