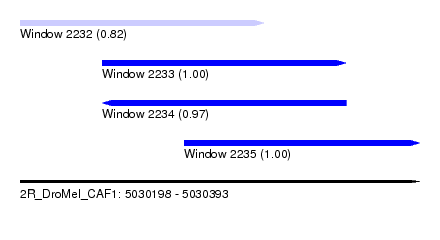
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,030,198 – 5,030,393 |
| Length | 195 |
| Max. P | 0.999849 |

| Location | 5,030,198 – 5,030,317 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5030198 119 + 20766785 GUAGAGAAACUGAAAAUAACUAAAUUAAGAGCGACUAAGUGUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG .((((((((........(((....(((((.(((......))).)))))..)))))))))))(((.(((.....))).)))..............(((((((((.....))))))))).. ( -27.10) >DroSec_CAF1 10255 119 + 1 GUAGAGAAACUAAAAAUAACUAAAUUAAGAGCGACUAAGUGUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG .((((((((........(((....(((((.(((......))).)))))..)))))))))))(((.(((.....))).)))..............(((((((((.....))))))))).. ( -27.10) >DroSim_CAF1 10281 119 + 1 GUAGAGAAACUGAAAAUAACUAAAUUAAGAGCGACUAAGUGUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG .((((((((........(((....(((((.(((......))).)))))..)))))))))))(((.(((.....))).)))..............(((((((((.....))))))))).. ( -27.10) >DroEre_CAF1 10179 116 + 1 GUAGAGAAACUAAGAAUAACUAAAUUAGGAGCGAAU---AGGGCUCGAGAGUUUCUCUCUGUGCUUGCAACUUGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG ..(((((((((........(((...)))((((....---...))))...)))))))))...(((.(((.....))).)))..............(((((((((.....))))))))).. ( -32.00) >DroYak_CAF1 10421 98 + 1 ------AAAUUAAGUGAAACUAAA---------------AGUUAUUAAGAGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG ------..................---------------((((....((((.....))))(((((.......))))).............))))(((((((((.....))))))))).. ( -21.20) >consensus GUAGAGAAACUAAAAAUAACUAAAUUAAGAGCGACUAAGUGUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUG ..((((((((........................................))))))))...(((.(((.....))).)))..............(((((((((.....))))))))).. (-18.92 = -19.44 + 0.52)

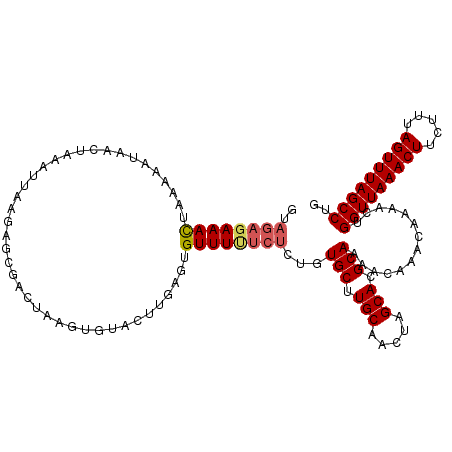
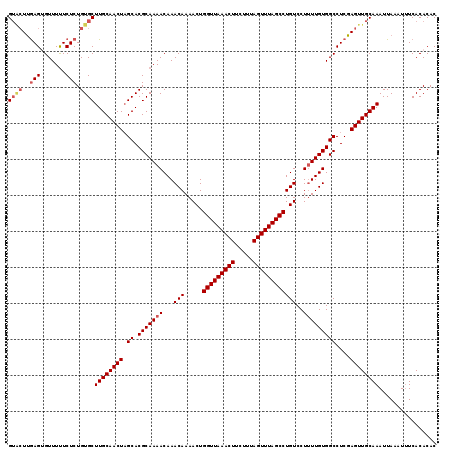
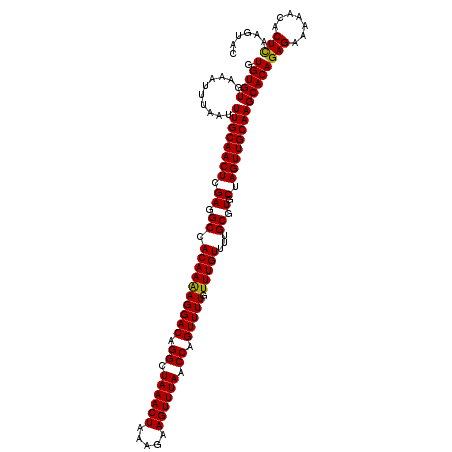
| Location | 5,030,238 – 5,030,357 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5030238 119 + 20766785 GUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACAC ((((..(((.......))).))))((((((((.((.(((((((...(((.....(((((((((.....))))))))))))..)))))))))....))))))))................ ( -33.70) >DroSec_CAF1 10295 118 + 1 GUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAU-UCACACAC ((((..(((.......))).))))((((((((.((.(((((((...(((.....(((((((((.....))))))))))))..)))))))))....)))))))).......-........ ( -33.70) >DroSim_CAF1 10321 119 + 1 GUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACAC ((((..(((.......))).))))((((((((.((.(((((((...(((.....(((((((((.....))))))))))))..)))))))))....))))))))................ ( -33.70) >DroEre_CAF1 10216 119 + 1 GGGCUCGAGAGUUUCUCUCUGUGCUUGCAACUUGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACAC .(((.((((((...))))).).)))(((((((((...((...(((((....((.(((((((((.....))))))))).))...))))).))..)))))))))................. ( -36.40) >DroYak_CAF1 10440 119 + 1 GUUAUUAAGAGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUCUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACAC .......((((.....))))....((((((((.((.(((((.....(((.....(((((((((.....))))))))))))....)))))))....))))))))................ ( -28.50) >consensus GUACUUGAGUGUUUUUCUCUGUGCUUGCAACUAGCACGCAAAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACAC ((((..(((.......))).))))((((((((.((.(((((((...(((.....(((((((((.....))))))))))))..)))))))))....))))))))................ (-29.76 = -30.64 + 0.88)

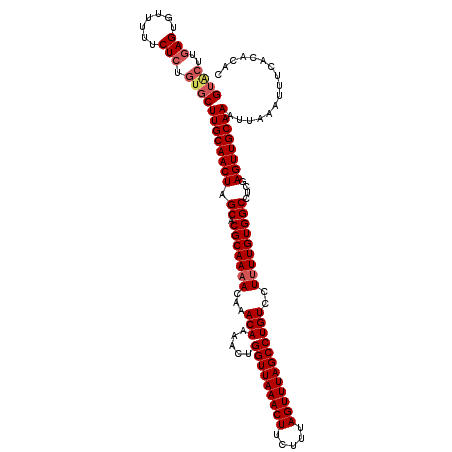
| Location | 5,030,238 – 5,030,357 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5030238 119 - 20766785 GUGUGUGAAAUUUAAUUUGCAACUCGAGGCCACAAAAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCUAGUUGCAAGCACAGAGAAAAACACUCAAGUAC (.((((....(((..(((((((((.((.((.((((((((((.((.((((((.....)))))).)).))))).)))))...)).).).)))))))))...))).....)))).)...... ( -28.60) >DroSec_CAF1 10295 118 - 1 GUGUGUGA-AUUUAAUUUGCAACUCGAGGCCACAAAAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCUAGUUGCAAGCACAGAGAAAAACACUCAAGUAC .(((((..-.......((((((((.((.((.((((((((((.((.((((((.....)))))).)).))))).)))))...)).).).)))))))))))))(((.......)))...... ( -28.60) >DroSim_CAF1 10321 119 - 1 GUGUGUGAAAUUUAAUUUGCAACUCGAGGCCACAAAAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCUAGUUGCAAGCACAGAGAAAAACACUCAAGUAC (.((((....(((..(((((((((.((.((.((((((((((.((.((((((.....)))))).)).))))).)))))...)).).).)))))))))...))).....)))).)...... ( -28.60) >DroEre_CAF1 10216 119 - 1 GUGUGUGAAAUUUAAUUUGCAACUCGAGGCCACAAAAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCAAGUUGCAAGCACAGAGAGAAACUCUCGAGCCC .(((((......(((((((((.(.((((((.((((...(((.((.((((((.....)))))).)).)))))))..)))))).))))))))))...)))))(((((...)))))...... ( -32.40) >DroYak_CAF1 10440 119 - 1 GUGUGUGAAAUUUAAUUUGCAACUCGAGGCCACAAGAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCUAGUUGCAAGCACAGAGAAAAACUCUUAAUAAC .(((((..........((((((((.((.((.((((((((((.((.((((((.....)))))).)).))))).)))))...)).).).)))))))))))))(((.....)))........ ( -28.40) >consensus GUGUGUGAAAUUUAAUUUGCAACUCGAGGCCACAAAAGGACAGGCUAAACUAAAGAAGUUUAACCAGUUUUGUUUGUUUUGCGUGCUAGUUGCAAGCACAGAGAAAAACACUCAAGUAC .(((((..........((((((((.((.((.((((((((((.((.((((((.....)))))).)).))))).)))))...)).).).)))))))))))))(((.......)))...... (-28.74 = -28.42 + -0.32)

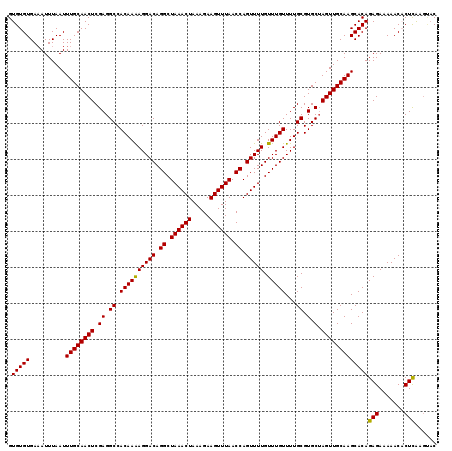
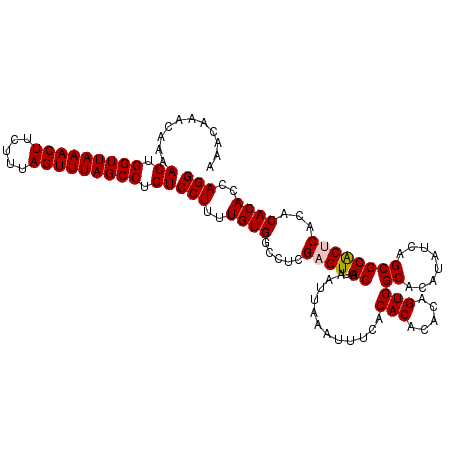
| Location | 5,030,278 – 5,030,393 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5030278 115 + 20766785 AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACACACAGUGGCACAUAUCAGCGCACUCACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((.....((((.((.............(((.....)))((........))))))))...))))..))) ( -26.70) >DroSec_CAF1 10335 114 + 1 AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAU-UCACACACACAGUGGCACAUAUCAGCGCACUCACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((.....((((.((.........-...(((.....)))((........))))))))...))))..))) ( -26.70) >DroSim_CAF1 10361 115 + 1 AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACACACAGUGGCACAUAUCAGCGCACUCACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((.....((((.((.............(((.....)))((........))))))))...))))..))) ( -26.70) >DroEre_CAF1 10256 115 + 1 AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACACACAGUGGCACACAUCCGCGCACACACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((((........)).....................(((((.(......).)).)))...))))..))) ( -23.20) >DroYak_CAF1 10480 115 + 1 AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUCUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACACACCGUGGCACAUAUCAGCGCGCACACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((((........)).....................(((((.(......).)).)))...))))..))) ( -23.30) >consensus AAACAAACAAAACUGGUUAAACUUCUUUAGUUUAGCCUGUCCUUUUGUGGCCUCGAGUUGCAAAUUAAAUUUCACACACACAGUGGCACAUAUCAGCGCACUCACACACACCAGG ...........((.(((((((((.....))))))))).))(((..((((.....((((.((.............(((.....)))((........))))))))...))))..))) (-24.58 = -24.82 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:10 2006