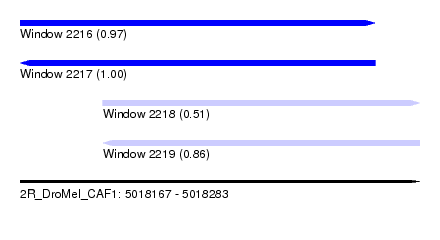
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,018,167 – 5,018,283 |
| Length | 116 |
| Max. P | 0.999209 |

| Location | 5,018,167 – 5,018,270 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -42.66 |
| Consensus MFE | -30.06 |
| Energy contribution | -31.18 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5018167 103 + 20766785 GGAGAAAGGCGAGAGGAG----------------AAAGGGAAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUAGCCUCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGA ((((((((.((.((((((----------------((.....(.((((((((.(((.(((..(((....)))..)))..))))))))))).)...))))).))))).))))))))..... ( -49.80) >DroSec_CAF1 72391 101 + 1 GGAGCAAGGCGAGAGUAG----------------AAAGGGAAGUGCGCUGGUGAGAAGACACGCAUUUGCGCACUG--CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGA ((((.((((((((((.((----------------((.(((.(((((((..(((.(......).)))..))))))).--)))((.....))....)))))))))..)))))))))..... ( -42.80) >DroSim_CAF1 75675 87 + 1 GGA--------------G----------------AAAGGGAAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGA (((--------------(----------------((((((.(((((((..(((.(......).)))..))))))).--)))...((.(((.(((....))).))))))))))))..... ( -36.70) >DroEre_CAF1 69389 110 + 1 GGAGAAAC------UGAGAAACG-GAGAAAGGGAGAAGGGAAGUCCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCUCCAGUGUGGUGAGUUCGCUCUCCGGCUUUCUCCGGGGA ........------.......((-((((((((((((.(.(((.((((((((......(((((((....)))...))--)).))))))....)).))).))))))..))))))))).... ( -43.40) >DroYak_CAF1 77833 117 + 1 GGAGAAGGGAGAUAGGAGAAAGGAGGGAAAGGGAAGAGGCAAGUCCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCACCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGA ..............((((((((...(((.((((((....((....((((((((.(.(((..(((....)))..)))--)))))))))...))..)))))).)))..))))))))..... ( -40.60) >consensus GGAGAAAGG_GA_AGGAG________________AAAGGGAAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUG__CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGA (((((((..............................(.......)(((((.((((.(((.(((.....(((((((......)))))))))).))).)))).))))))))))))..... (-30.06 = -31.18 + 1.12)

| Location | 5,018,167 – 5,018,270 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.86 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5018167 103 - 20766785 UCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGAGGCUAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUUCCCUUU----------------CUCCUCUCGCCUUUCUCC .....((((((((.(((((.(((((((((......))))(((..(((((((...((.(((......))))))))))))..)))))----------------)))))).)).)))))))) ( -42.70) >DroSec_CAF1 72391 101 - 1 UCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG--CAGUGCGCAAAUGCGUGUCUUCUCACCAGCGCACUUCCCUUU----------------CUACUCUCGCCUUGCUCC .....((((.(((.((((((((((.....((.....))(((--.(((((((...((.(((......)))))))))))).))))))----------------)).))).)).))).)))) ( -38.70) >DroSim_CAF1 75675 87 - 1 UCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUUCCCUUU----------------C--------------UCC ...((((......)))).((((((.....((.....))(((--.(((((((...((.(((......)))))))))))).))))))----------------)--------------)). ( -32.80) >DroEre_CAF1 69389 110 - 1 UCCCCGGAGAAAGCCGGAGAGCGAACUCACCACACUGGAGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGGACUUCCCUUCUCCCUUUCUC-CGUUUCUCA------GUUUCUCC ....(((((((((..((((((.(((.((....(.(((((((--..(..(((....)))..)..)))..)))).))).))).).))))))))))))-)).......------........ ( -36.40) >DroYak_CAF1 77833 117 - 1 UCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGUGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGGACUUGCCUCUUCCCUUUCCCUCCUUUCUCCUAUCUCCCUUCUCC .....((((((((..((((((.(((.........(((((((--..(..(((....)))..)....))))))).((.....))..))).))))))...)))))))).............. ( -39.40) >consensus UCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG__CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUUCCCUUU________________CUCCU_UC_CCUUUCUCC .....((((((((..((.(((....))).)).(.(((((((....(..(((....)))..)...))).)))).).....................................)))))))) (-24.18 = -25.86 + 1.68)

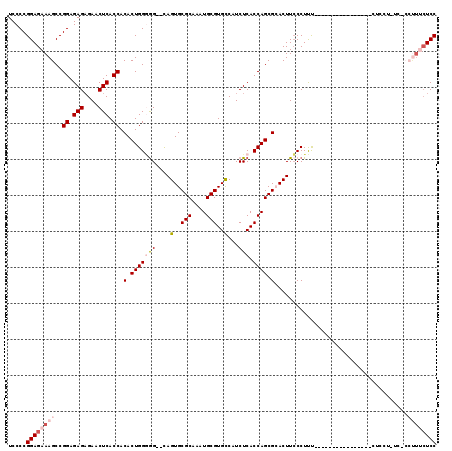
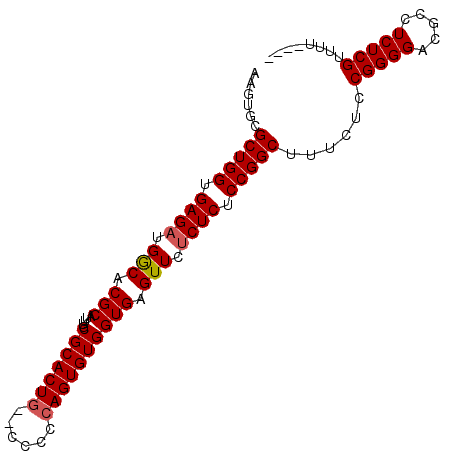
| Location | 5,018,191 – 5,018,283 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5018191 92 + 20766785 AAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUAGCCUCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGACGCCUCUCGUUUU---- .(.((((((((.(((.(((..(((....)))..)))..))))))))))).)(((.....(((((((......))))))).....))).....---- ( -35.00) >DroSec_CAF1 72415 90 + 1 AAGUGCGCUGGUGAGAAGACACGCAUUUGCGCACUG--CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGACGCCUCUCGUUUU---- .((.(((((((.((((..((.(((.....(((((((--....)))))))))).))..)))).))))).......(....)))))........---- ( -32.40) >DroSim_CAF1 75685 90 + 1 AAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGACGCCUCUCGUUUU---- .((.(((((((.((((..((.(((.....(((((((--....)))))))))).))..)))).))))).......(....)))))........---- ( -31.80) >DroEre_CAF1 69422 94 + 1 AAGUCCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCUCCAGUGUGGUGAGUUCGCUCUCCGGCUUUCUCCGGGGACGCCUCUCGGUUUUUUA ..((((.((((.((((.(.(..((....))((((((--....))))))((.(((....))).))).))))).))))))))(((....)))...... ( -34.20) >DroYak_CAF1 77873 90 + 1 AAGUCCGCUGGUGAGAUGGCACGCAUUUGCGCACUG--CCACCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGACGCCUCUCGUUUU---- ..(((((((((.((((..((.(((.....(((((((--....)))))))))).))..)))).)))))...(....)))))............---- ( -32.20) >consensus AAGUGCGCUGGUGAGAUGGCACGCAUUUGCGCACUG__CCCCCAGUGUGGUGAGUUCUCUCUCCGGCUUUCUCCGGGGACGCCUCUCGUUUU____ ......(((((.((((.(((.(((.....(((((((......)))))))))).))).)))).)))))......(((((.....)))))........ (-29.10 = -29.34 + 0.24)

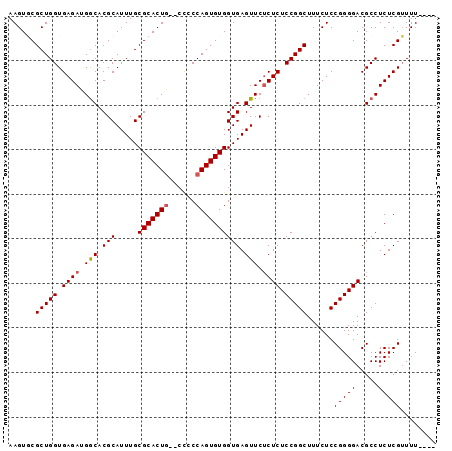
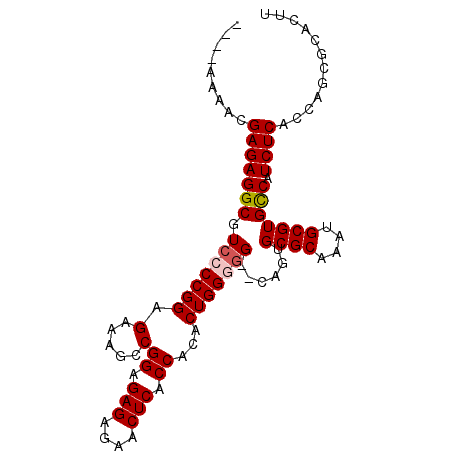
| Location | 5,018,191 – 5,018,283 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.40 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5018191 92 - 20766785 ----AAAACGAGAGGCGUCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGAGGCUAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUU ----.....(((((((.((((.((......))((.(((....))).)).....)))).))).(..(((....)))..)..))))............ ( -31.10) >DroSec_CAF1 72415 90 - 1 ----AAAACGAGAGGCGUCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG--CAGUGCGCAAAUGCGUGUCUUCUCACCAGCGCACUU ----....(....)..((((((((.(.....)((.(((....))).))...)))))))--)(((((((...((.(((......)))))))))))). ( -33.30) >DroSim_CAF1 75685 90 - 1 ----AAAACGAGAGGCGUCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUU ----.....(((((((((((((((.(.....)((.(((....))).))...)))))))--)...((((....))))))).))))............ ( -35.00) >DroEre_CAF1 69422 94 - 1 UAAAAAACCGAGAGGCGUCCCCGGAGAAAGCCGGAGAGCGAACUCACCACACUGGAGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGGACUU .......(((.(((.((..(((((......)))).)..))..)))......(((((((--..(..(((....)))..)..)))..))))))).... ( -31.40) >DroYak_CAF1 77873 90 - 1 ----AAAACGAGAGGCGUCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGUGG--CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGGACUU ----....(....)..((((((((......)))).(((....)))......(((((((--..(..(((....)))..)....))))))).)))).. ( -31.50) >consensus ____AAAACGAGAGGCGUCCCCGGAGAAAGCCGGAGAGAGAACUCACCACACUGGGGG__CAGUGCGCAAAUGCGUGCCAUCUCACCAGCGCACUU .........(((((((.(((((((.(.....)((.(((....))).))...)))))))......((((....))))))).))))............ (-25.78 = -26.22 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:51 2006