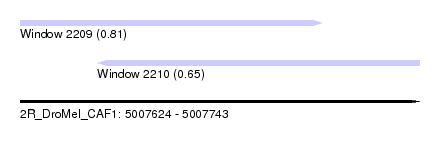
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 5,007,624 – 5,007,743 |
| Length | 119 |
| Max. P | 0.805037 |

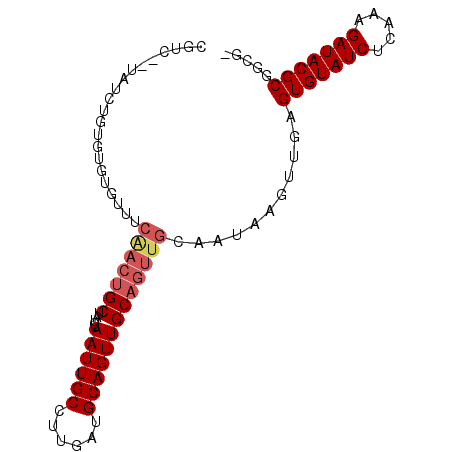
| Location | 5,007,624 – 5,007,714 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5007624 90 + 20766785 UGUC--UCUGUGUGUGUGCUUCAACUGCUAUCUAAUUGCCUUGAUGCAGUUGCAGUUGCAAUAAGUUGAGUGUAUCUCAAAGAUACGCGAUG- .(((--.((...((((.(((.(((((((.(((..........)))))))))).)))))))...))....(((((((.....)))))))))).- ( -26.20) >DroPse_CAF1 63027 93 + 1 CGUCCGUAUCUGUGGCUGUUUCGACUGCUAUCUAAUUGCCCCGAUGCAGUUGCAGUUGCAAUGAGUGGAGUGUAUCUUUAAGAUACGCGGCCC .....(((.(((..(((((.(((...((.........))..))).)))))..))).))).......((.(((((((.....)))))))..)). ( -32.30) >DroSim_CAF1 64143 90 + 1 UGUC--UCUGUGUGUGUGCUUCAACUGCUAUCUAAUUGCCUUGAUGCAGUUGCAGUUGCAAUAAGUUGAGUGUAUCUCAAAGAUACGCGGCG- .(((--.((...((((.(((.(((((((.(((..........)))))))))).)))))))...))....(((((((.....)))))))))).- ( -27.70) >DroYak_CAF1 66831 90 + 1 GCUC--UGUGUGUGUGUGUUUCAACUGCUAUCUAAUUGCCUUGAUGCAGUUGCAGUUGCAAUAAGUUGAGUGUAUCUCAAAGAUACGCGCUG- ....--.((((((((.((((.(((((((.....((((((......))))))))))))).))))..(((((.....)))))..))))))))..- ( -28.30) >DroAna_CAF1 62642 77 + 1 GCCA--GGUCUG-------------CGCUAUCUAAUUGCCUUGAUGCAGUUGCAGUUGCAAUAAGUUGAGUGUAUCUUUAAGAUACGCGACU- ....--((((.(-------------((.(((((((.(((((..((...(((((....)))))..))..)).)))...)).))))))))))))- ( -22.70) >DroPer_CAF1 63217 93 + 1 CGUCCGUAUCUGUGGCUGUUUCGACUGCUAUCUAAUUGCCCCGAUGCAGUUGCAGUUGCAAUGAGUGGAGUGUAUCUUUAAGAUACGCGGCCC .....(((.(((..(((((.(((...((.........))..))).)))))..))).))).......((.(((((((.....)))))))..)). ( -32.30) >consensus CGUC__UAUCUGUGUGUGUUUCAACUGCUAUCUAAUUGCCUUGAUGCAGUUGCAGUUGCAAUAAGUUGAGUGUAUCUCAAAGAUACGCGGCG_ .....................(((((((.....((((((......)))))))))))))...........(((((((.....)))))))..... (-19.00 = -19.67 + 0.67)

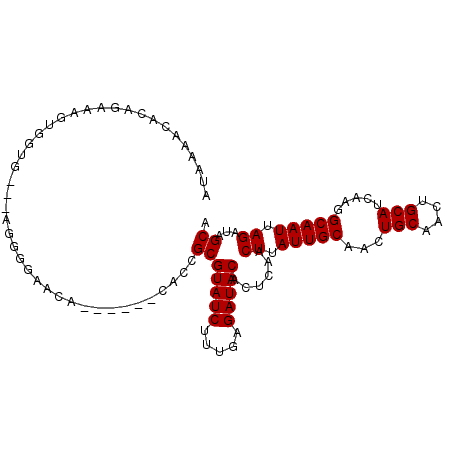
| Location | 5,007,647 – 5,007,743 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 5007647 96 - 20766785 AUAAAACACAGAAAUUGGUU---GGGGGAACA------CAUCGCGUAUCUUUGAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCA ................((((---(((.((...------..))..(((((.....))))).))))))).(((((...(((....))).....)))))......... ( -17.90) >DroSec_CAF1 61813 96 - 1 AUAAAACACAGAAAGUGGUG---GGGGGAGCA------CACCGCGUAUCUUUGAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCA ..........((..((((((---..(....).------))))))(((((.....)))))..))..((.(((((...(((....))).....))))).))...... ( -23.70) >DroSim_CAF1 64166 96 - 1 AUAAGACACAGAAAGUGGUG---GGGGGAACA------CGCCGCGUAUCUUUGAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCA ..........((..((((((---..(....).------))))))(((((.....)))))..))..((.(((((...(((....))).....))))).))...... ( -24.30) >DroYak_CAF1 66854 96 - 1 AUAAAACACAGAAAGUGGUG---AGGGGAACA------CAGCGCGUAUCUUUGAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCA ......(((.....)))((.---..(....).------..))(((((((.....)))))......((.(((((...(((....))).....))))).))...)). ( -20.00) >DroAna_CAF1 62652 88 - 1 -----AGGCAGAAAACGGCC---AACUAA---------AGUCGCGUAUCUUAAAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCG -----.(((........)))---......---------...((((((((.....)))))......((.(((((...(((....))).....))))).))...))) ( -20.90) >DroPer_CAF1 63242 96 - 1 ---------AGAGAGUGGGGGAGAGAGGAACCGAGUGGGGCCGCGUAUCUUAAAGAUACACUCCACUCAUUGCAACUGCAACUGCAUCGGGGCAAUUAGAUAGCA ---------...((((((((....(.((..((....))..)).)(((((.....))))).))))))))(((((..((((....)))..)..)))))......... ( -33.30) >consensus AUAAAACACAGAAAGUGGUG___AGGGGAACA______CACCGCGUAUCUUUGAGAUACACUCAACUUAUUGCAACUGCAACUGCAUCAAGGCAAUUAGAUAGCA ..........................................(((((((.....)))))......((.(((((...(((....))).....))))).))...)). (-16.02 = -16.02 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:43 2006