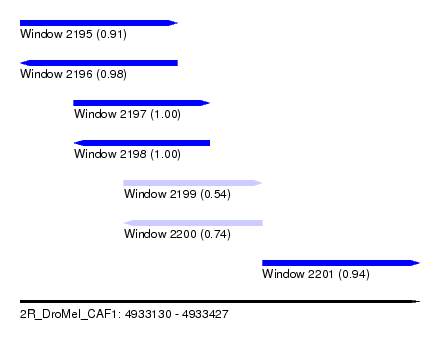
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,933,130 – 4,933,427 |
| Length | 297 |
| Max. P | 0.999635 |

| Location | 4,933,130 – 4,933,247 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -24.56 |
| Energy contribution | -26.62 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933130 117 + 20766785 CGAUGGACAUAAUCCCAUCGAAAUUGCUUAUCAACUGCGGACAAACGAACGAGCACUCA---UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAG ((((((........))))))....(((((........((......))...)))))....---...((((((((.((((....)))))))))))).........((((((....)))))). ( -37.40) >DroSim_CAF1 1393 112 + 1 CGAUGGGCCUAAUCCCACCGAAAUUGCUUAUCAACUGCGGA--------CGAGCACUCAUCAUCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAG ((.((((......)))).))....((((..((.......))--------..))))..........((((((((.((((....)))))))))))).........((((((....)))))). ( -37.50) >DroYak_CAF1 1383 117 + 1 AGAUGGGCCUAAUCCCACUGAAAUUGCUUAUCAACUGCGGACAAACGAAUGGGCACUCA---UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUCGCAAUGGAAAUUGCAG .((((((((((.((....((...((((.........)))).))...)).))))).).))---)).((((((((.((((....))))))))))))..........(((((....))))).. ( -37.90) >DroAna_CAF1 4214 113 + 1 CGAUGGGCCUAAUCCCCCUGAAAUUGCUUAUCAACUCCGCGUAGAGA----GAUACUCA---UCUUCUAAAUGACCCGUUGUUGGCCAUUUGGCUAAGUACAUUGCAAUGAAAAUUGCUG (((((((.....((.....(((..((..((((..(((......))).----))))..))---..))).....)))))))))((((((....)))))).......(((((....))))).. ( -26.90) >consensus CGAUGGGCCUAAUCCCACCGAAAUUGCUUAUCAACUGCGGACAAACG__CGAGCACUCA___UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAG .(((((((....((.....))....))))))).................................((((((((.((((....))))))))))))..........(((((....))))).. (-24.56 = -26.62 + 2.06)

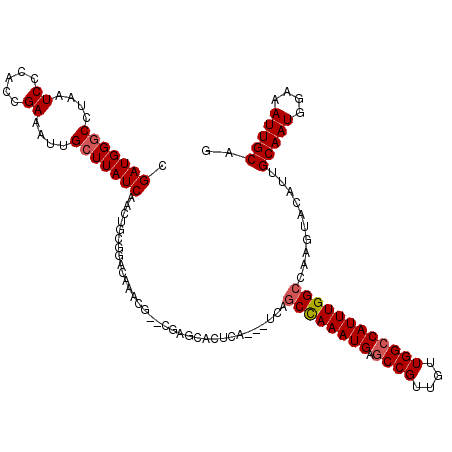
| Location | 4,933,130 – 4,933,247 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -28.48 |
| Energy contribution | -29.48 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933130 117 - 20766785 CUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA---UGAGUGCUCGUUCGUUUGUCCGCAGUUGAUAAGCAAUUUCGAUGGGAUUAUGUCCAUCG .((((((....))))))..((((((((((((((((((......))).)))))))))..---.))))))......(((((((.......))))))).....(((((((......))))))) ( -42.30) >DroSim_CAF1 1393 112 - 1 CUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGAUGAUGAGUGCUCG--------UCCGCAGUUGAUAAGCAAUUUCGGUGGGAUUAGGCCCAUCG .(((....((.(((((.......(..(((((((((((......))).))))))))..).(((((....)))--------)).))))).))...)))....(((((((......))))))) ( -43.20) >DroYak_CAF1 1383 117 - 1 CUGCAAUUUCCAUUGCGAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA---UGAGUGCCCAUUCGUUUGUCCGCAGUUGAUAAGCAAUUUCAGUGGGAUUAGGCCCAUCU ..((...((((((((.(((((((((((((((((((((......))).)))))))))..---.))))))......(((((((.......))))))).))).))))))))....))...... ( -41.70) >DroAna_CAF1 4214 113 - 1 CAGCAAUUUUCAUUGCAAUGUACUUAGCCAAAUGGCCAACAACGGGUCAUUUAGAAGA---UGAGUAUC----UCUCUACGCGGAGUUGAUAAGCAAUUUCAGGGGGAUUAGGCCCAUCG ..(((((....))))).(((..(((((((((((((((.......)))))))).((((.---((..((((----.((((....))))..))))..)).))))....)).)))))..))).. ( -30.60) >consensus CUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA___UGAGUGCCCG__CGUUUGUCCGCAGUUGAUAAGCAAUUUCAGUGGGAUUAGGCCCAUCG ..(((((....)))))...((((((((((((((((((......))).)))))))))......))))))..............(.(((((.....))))).)((((((......)))))). (-28.48 = -29.48 + 1.00)

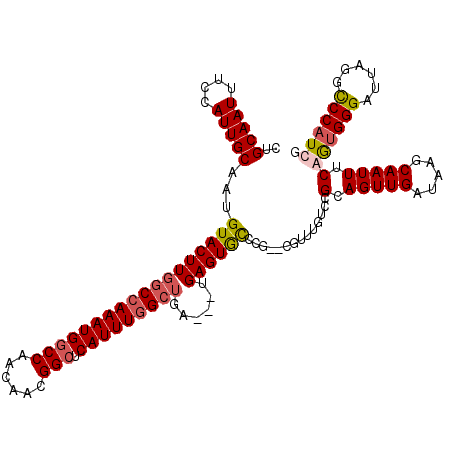

| Location | 4,933,170 – 4,933,271 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933170 101 + 20766785 ACAAACGAACGAGCACUCA---UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUCCCCG ..........(((((((..---...((((((((.((((....))))))))))))(((((.(((((((((....))))))))------)...)))))..-))---------).)))).... ( -39.10) >DroSim_CAF1 1433 96 + 1 A--------CGAGCACUCAUCAUCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUCCCCG .--------.(((((((........((((((((.((((....))))))))))))(((((.(((((((((....))))))))------)...)))))..-))---------).)))).... ( -39.10) >DroYak_CAF1 1423 101 + 1 ACAAACGAAUGGGCACUCA---UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUCGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUUCCCG .....(((...((((....---...((((((((.((((....))))))))))))...((.(((.(((((....))))).))------)))....))))-..---------)))....... ( -34.60) >DroAna_CAF1 4254 113 + 1 GUAGAGA----GAUACUCA---UCUUCUAAAUGACCCGUUGUUGGCCAUUUGGCUAAGUACAUUGCAAUGAAAAUUGCUGCUGGUUCUGCAAUUUUCUCAGUUUUUUUUACCCCUUCCCG (((((((----((.(((..---...............((..((((((....))))))..))........(((((((((..........)))))))))..))))))))))))......... ( -24.90) >consensus ACAAACG__CGAGCACUCA___UCAGCCAAAUGAGCCGUUGUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU______GGCAAUUUGCU_AG_________UCGCUCCCCG ..........((((...........((((((((.((((....))))))))))))...............(.(((((((..........))))))).)...............)))).... (-22.36 = -23.05 + 0.69)

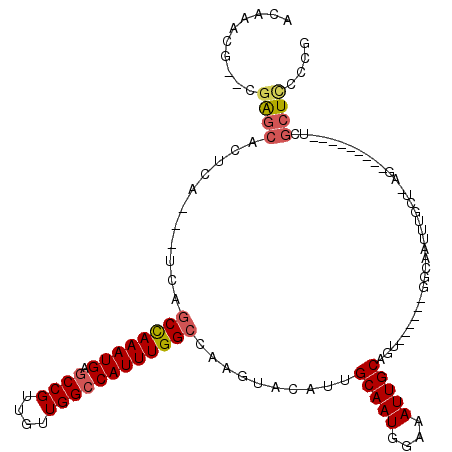
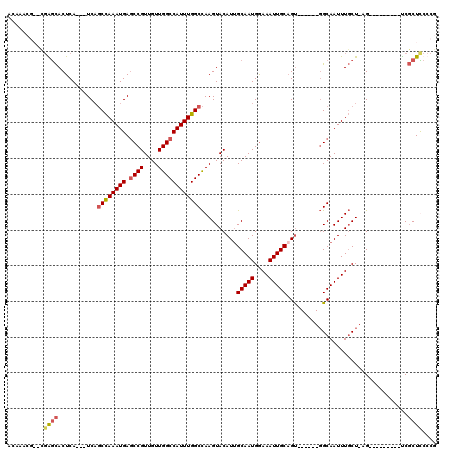
| Location | 4,933,170 – 4,933,271 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933170 101 - 20766785 CGGGGAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA---UGAGUGCUCGUUCGUUUGU ....((((((---------..-.(((((((((.------...))))).....))))...((((((((((((((((((......))).)))))))))..---.))))))))))))...... ( -37.80) >DroSim_CAF1 1433 96 - 1 CGGGGAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGAUGAUGAGUGCUCG--------U ....((((.(---------((-..((......(------(.((((((....)))))).))...(..(((((((((((......))).))))))))..)))...))))))).--------. ( -34.70) >DroYak_CAF1 1423 101 - 1 CGGGAAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCGAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA---UGAGUGCCCAUUCGUUUGU .(((((((..---------..-.))...((((.------...)))))))))...((((.((((((((((((((((((......))).)))))))))..---.))))))....)))).... ( -35.80) >DroAna_CAF1 4254 113 - 1 CGGGAAGGGGUAAAAAAAACUGAGAAAAUUGCAGAACCAGCAGCAAUUUUCAUUGCAAUGUACUUAGCCAAAUGGCCAACAACGGGUCAUUUAGAAGA---UGAGUAUC----UCUCUAC ..(((.(((..............(((((((((..........)))))))))........(((((((.(.((((((((.......)))))))).)....---))))))))----))))).. ( -27.30) >consensus CGGGAAGCGA_________CU_AGCAAAUUGCC______ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAACAACGGCUCAUUUGGCUGA___UGAGUGCCCG__CGUUUGU .......................(.(((((((..........))))))).)........((((((((((((((((((......))).)))))))))......))))))............ (-22.00 = -22.62 + 0.62)

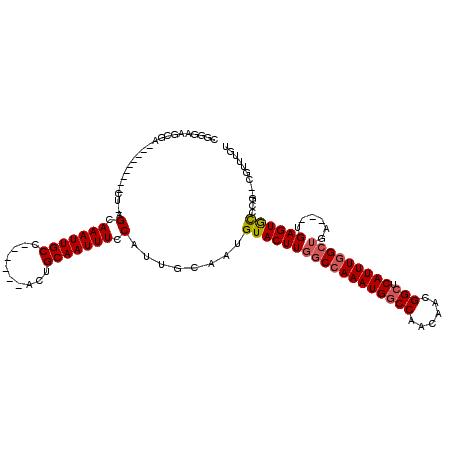
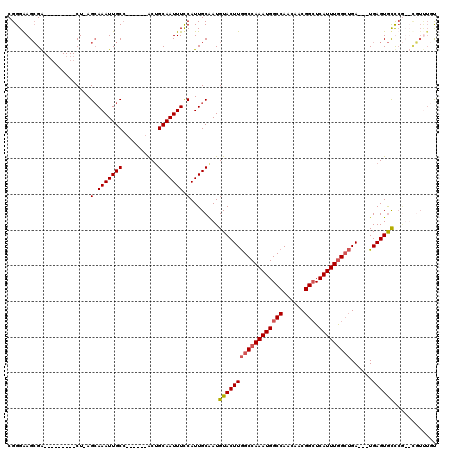
| Location | 4,933,207 – 4,933,310 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -16.48 |
| Energy contribution | -15.91 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933207 103 + 20766785 GUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUCCCCGCAAAAUCUCUGCCCCGCACUCGU-UUUAAUUGAAUAAACA .((((((....))))))...(((((((((....))))))))------).(((((.((.-((---------(.((.....(((.......)))...))))).))-...)))))........ ( -30.30) >DroSim_CAF1 1465 103 + 1 GUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUCCCCGCAAAAUCUCUGCCCCGCACUCGU-UUUAAUUGAAUAAACA .((((((....))))))...(((((((((....))))))))------).(((((.((.-((---------(.((.....(((.......)))...))))).))-...)))))........ ( -30.30) >DroYak_CAF1 1460 102 + 1 GUUGGCCAUUUGGCCAAGUACAUCGCAAUGGAAAUUGCAGU------GGCAAUUUGCU-AG---------UCGCUUCCCGCCAAAUCUCUGCCCC-CACUCUU-UUUAAUUGAAUAAACA ...(((.(((((((.((((((...(((((....)))))..(------(((.....)))-))---------).))))...)))))))....)))..-.......-................ ( -24.00) >DroAna_CAF1 4287 120 + 1 GUUGGCCAUUUGGCUAAGUACAUUGCAAUGAAAAUUGCUGCUGGUUCUGCAAUUUUCUCAGUUUUUUUUACCCCUUCCCGCCAAAUCUCUGGCCCGCACUCUUUUUUAAUUGAAUAAACA ((.(((((((((((...((.....))...(((((((((..........)))))))))......................))))))....))))).))....................... ( -26.70) >consensus GUUGGCCAUUUGGCCAAGUACAUUGCAAUGGAAAUUGCAGU______GGCAAUUUGCU_AG_________UCGCUCCCCGCAAAAUCUCUGCCCCGCACUCGU_UUUAAUUGAAUAAACA .((((((....))))))((......(((((.(((((((..........))))))).)...............((................))................)))).....)). (-16.48 = -15.91 + -0.56)

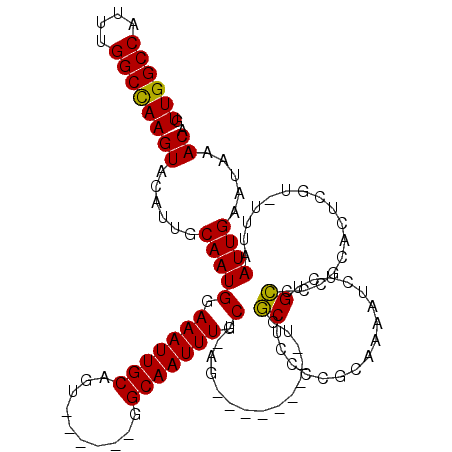
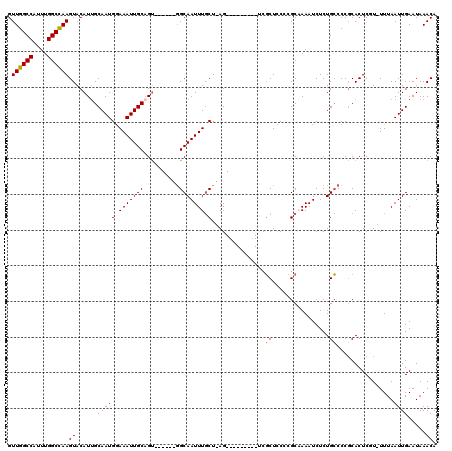
| Location | 4,933,207 – 4,933,310 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933207 103 - 20766785 UGUUUAUUCAAUUAAA-ACGAGUGCGGGGCAGAGAUUUUGCGGGGAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAAC ................-..(((((((..((((.((..((((((.(.((((---------..-......)))))------.))))))..))..))))..)))))))((((....))))... ( -33.70) >DroSim_CAF1 1465 103 - 1 UGUUUAUUCAAUUAAA-ACGAGUGCGGGGCAGAGAUUUUGCGGGGAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAAC ................-..(((((((..((((.((..((((((.(.((((---------..-......)))))------.))))))..))..))))..)))))))((((....))))... ( -33.70) >DroYak_CAF1 1460 102 - 1 UGUUUAUUCAAUUAAA-AAGAGUG-GGGGCAGAGAUUUGGCGGGAAGCGA---------CU-AGCAAAUUGCC------ACUGCAAUUUCCAUUGCGAUGUACUUGGCCAAAUGGCCAAC ................-...((((-((.((((.....((((((...((..---------..-.))...)))))------)))))....)))))).........((((((....)))))). ( -28.10) >DroAna_CAF1 4287 120 - 1 UGUUUAUUCAAUUAAAAAAGAGUGCGGGCCAGAGAUUUGGCGGGAAGGGGUAAAAAAAACUGAGAAAAUUGCAGAACCAGCAGCAAUUUUCAUUGCAAUGUACUUAGCCAAAUGGCCAAC ....(((((..........)))))..(((((....((((((...(((.(((.......)))..(((((((((..........)))))))))...........))).)))))))))))... ( -28.40) >consensus UGUUUAUUCAAUUAAA_AAGAGUGCGGGGCAGAGAUUUGGCGGGAAGCGA_________CU_AGCAAAUUGCC______ACUGCAAUUUCCAUUGCAAUGUACUUGGCCAAAUGGCCAAC ...................(((((((..((((.......((.....))...............(.(((((((..........))))))).).))))..)))))))((((....))))... (-20.80 = -21.80 + 1.00)

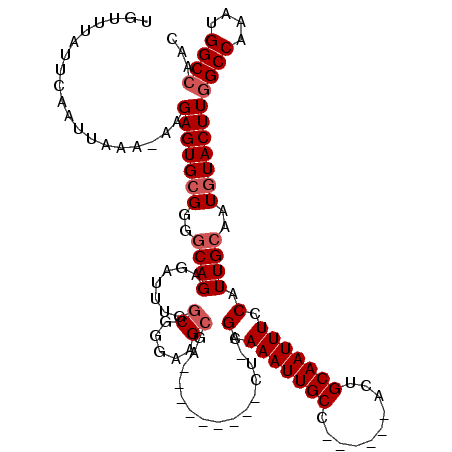
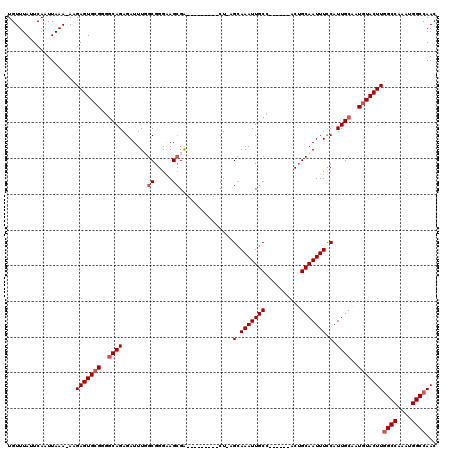
| Location | 4,933,310 – 4,933,427 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -21.52 |
| Energy contribution | -25.27 |
| Covariance contribution | 3.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4933310 117 + 20766785 CAGCGGUUAGUGUGUUUACAUAUUGAGUGCAUCAGCAGCAGUUGCAACAUGCGGCGUGCAGCGCGCAACGUGCUACGUGCAACAAUUGCA---GCUGCUGCCGGAGACAUGGCCAGCGAU ..(((.((((((((....)))))))).)))(((.(((((((((((((..((..(((((.((((((...)))))).)))))..)).)))))---)))))))).(....).........))) ( -51.60) >DroSim_CAF1 1568 103 + 1 CAGCGGUUAGUGUGUUUACAUAUUGAGUGAAUCAGCAGCAGUUGCAACAUGCGGCGUGCA--------------ACGUGCAACAAUUGCA---GCUGCUGCCGGCGACAUGGCCAGCGAU ...((.((((((((....)))))))).)).(((.(((((((((((((..((((.((....--------------.))))))....)))))---)))))))).(((......)))...))) ( -41.50) >DroYak_CAF1 1562 110 + 1 CAGCGGUUAGUGUGUUUACAUAUUGAGUGCAUCAGCAGCAGUUGCAACAUGCUGCGUGCA-------ACGUGCAACGUGCCACAAUUGCA---GCUGCUGCCGGCGACAUGGCCACCGAU ..(((.((((((((....)))))))).)))(((.(((((((((((((..((..((((((.-------....)).))))..))...)))))---)))))))).(((......)))...))) ( -47.70) >DroAna_CAF1 4407 94 + 1 CAGCGGUUAGUGUAUUUACAUUUUGAGUGCAUCC-----AUUG-------GCU--------------ACGUGCAACGUGCAACAGUUGCAGCAGCUGCUGCCGGCGACAUGGCCACCGAU ...((((..(((((((((.....)))))))))..-----...(-------(((--------------(.((((..((.(((.((((((...)))))).))))))).)).))))))))).. ( -31.80) >consensus CAGCGGUUAGUGUGUUUACAUAUUGAGUGCAUCAGCAGCAGUUGCAACAUGCGGCGUGCA_______ACGUGCAACGUGCAACAAUUGCA___GCUGCUGCCGGCGACAUGGCCACCGAU ..(((.((((((((....)))))))).)))..(((((((.(((((.....)))))......................((((.....))))...)))))))..(((......)))...... (-21.52 = -25.27 + 3.75)

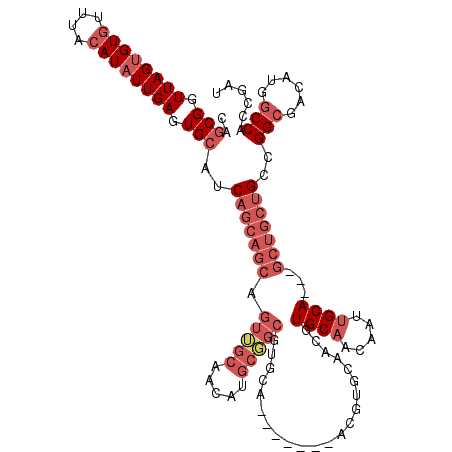
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:34 2006